
Sistotremastrum niveocremeum HHB9708
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Trechisporales; Trechisporales incertae sedis; Sistotremastrum; Sistotremastrum niveocremeum
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
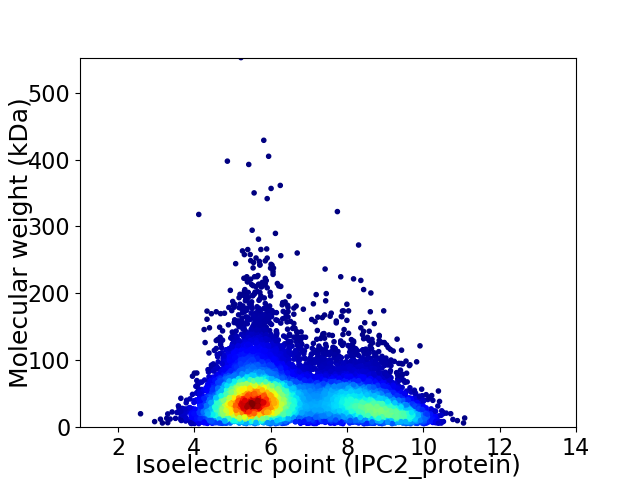
Virtual 2D-PAGE plot for 13037 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A164RVU1|A0A164RVU1_9AGAM Uncharacterized protein OS=Sistotremastrum niveocremeum HHB9708 OX=1314777 GN=SISNIDRAFT_468170 PE=4 SV=1
VV1 pKa = 7.84DD2 pKa = 4.52DD3 pKa = 5.55ASDD6 pKa = 3.54VTYY9 pKa = 7.56TTTMNIGGRR18 pKa = 11.84DD19 pKa = 3.28ISVLIDD25 pKa = 3.37TGSSDD30 pKa = 3.7LWATGQIPNSTPTNFTADD48 pKa = 3.16VTYY51 pKa = 10.8AQGDD55 pKa = 3.41AGGYY59 pKa = 9.64ISIAPVQFAGYY70 pKa = 7.27TIPDD74 pKa = 3.23QAYY77 pKa = 9.01INVTSGDD84 pKa = 3.93PPSVDD89 pKa = 3.15GLIGLGPYY97 pKa = 8.83MGSTIRR103 pKa = 11.84TTVGSPSADD112 pKa = 2.91PVLNRR117 pKa = 11.84IFAQNTSTPNFLSFILGRR135 pKa = 11.84SSGPSQPFSGEE146 pKa = 3.91LTVGEE151 pKa = 4.46VIQGFEE157 pKa = 4.35KK158 pKa = 10.53ISGEE162 pKa = 4.25VKK164 pKa = 10.85NEE166 pKa = 3.85VQVLPTSDD174 pKa = 5.06SGDD177 pKa = 3.55QHH179 pKa = 6.37WALSLDD185 pKa = 3.98TFTGPDD191 pKa = 3.5GKK193 pKa = 10.53AIPYY197 pKa = 6.68TTTVPNEE204 pKa = 4.24SKK206 pKa = 10.47PVAVLDD212 pKa = 4.11SGFSLPQIPGNISDD226 pKa = 5.26AIYY229 pKa = 10.78GRR231 pKa = 11.84VQGAQWLTTSPVGPVWQIPCEE252 pKa = 3.97QEE254 pKa = 4.28LNISVTIGGKK264 pKa = 9.69VYY266 pKa = 9.92PIHH269 pKa = 6.73PLDD272 pKa = 3.58VSLEE276 pKa = 3.87ITINPDD282 pKa = 2.62IPNICVGAFQPSTIGFSNAAQIFDD306 pKa = 4.09MVLGMSFLRR315 pKa = 11.84NTYY318 pKa = 7.84TLINFGDD325 pKa = 4.0FVDD328 pKa = 4.3VNSSVTADD336 pKa = 4.01PYY338 pKa = 10.89IQLLSLTDD346 pKa = 3.89PGQAHH351 pKa = 6.28QDD353 pKa = 3.73FVSTRR358 pKa = 11.84LNNVDD363 pKa = 3.77TTSDD367 pKa = 3.45SQFALLPASSQQSSS381 pKa = 3.05
VV1 pKa = 7.84DD2 pKa = 4.52DD3 pKa = 5.55ASDD6 pKa = 3.54VTYY9 pKa = 7.56TTTMNIGGRR18 pKa = 11.84DD19 pKa = 3.28ISVLIDD25 pKa = 3.37TGSSDD30 pKa = 3.7LWATGQIPNSTPTNFTADD48 pKa = 3.16VTYY51 pKa = 10.8AQGDD55 pKa = 3.41AGGYY59 pKa = 9.64ISIAPVQFAGYY70 pKa = 7.27TIPDD74 pKa = 3.23QAYY77 pKa = 9.01INVTSGDD84 pKa = 3.93PPSVDD89 pKa = 3.15GLIGLGPYY97 pKa = 8.83MGSTIRR103 pKa = 11.84TTVGSPSADD112 pKa = 2.91PVLNRR117 pKa = 11.84IFAQNTSTPNFLSFILGRR135 pKa = 11.84SSGPSQPFSGEE146 pKa = 3.91LTVGEE151 pKa = 4.46VIQGFEE157 pKa = 4.35KK158 pKa = 10.53ISGEE162 pKa = 4.25VKK164 pKa = 10.85NEE166 pKa = 3.85VQVLPTSDD174 pKa = 5.06SGDD177 pKa = 3.55QHH179 pKa = 6.37WALSLDD185 pKa = 3.98TFTGPDD191 pKa = 3.5GKK193 pKa = 10.53AIPYY197 pKa = 6.68TTTVPNEE204 pKa = 4.24SKK206 pKa = 10.47PVAVLDD212 pKa = 4.11SGFSLPQIPGNISDD226 pKa = 5.26AIYY229 pKa = 10.78GRR231 pKa = 11.84VQGAQWLTTSPVGPVWQIPCEE252 pKa = 3.97QEE254 pKa = 4.28LNISVTIGGKK264 pKa = 9.69VYY266 pKa = 9.92PIHH269 pKa = 6.73PLDD272 pKa = 3.58VSLEE276 pKa = 3.87ITINPDD282 pKa = 2.62IPNICVGAFQPSTIGFSNAAQIFDD306 pKa = 4.09MVLGMSFLRR315 pKa = 11.84NTYY318 pKa = 7.84TLINFGDD325 pKa = 4.0FVDD328 pKa = 4.3VNSSVTADD336 pKa = 4.01PYY338 pKa = 10.89IQLLSLTDD346 pKa = 3.89PGQAHH351 pKa = 6.28QDD353 pKa = 3.73FVSTRR358 pKa = 11.84LNNVDD363 pKa = 3.77TTSDD367 pKa = 3.45SQFALLPASSQQSSS381 pKa = 3.05
Molecular weight: 40.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A165A9D2|A0A165A9D2_9AGAM Cytochrome P450 OS=Sistotremastrum niveocremeum HHB9708 OX=1314777 GN=SISNIDRAFT_481369 PE=3 SV=1
MM1 pKa = 7.39PRR3 pKa = 11.84IPFSLSRR10 pKa = 11.84LLARR14 pKa = 11.84PLPIVQRR21 pKa = 11.84QPLVNRR27 pKa = 11.84SVSLTTHH34 pKa = 7.08LSSSIPRR41 pKa = 11.84LSSSPILRR49 pKa = 11.84AVSLLRR55 pKa = 11.84SSISLLPTTPTASHH69 pKa = 7.22LLQVRR74 pKa = 11.84FNQRR78 pKa = 11.84GVDD81 pKa = 3.75YY82 pKa = 10.92QPSQRR87 pKa = 11.84KK88 pKa = 8.81RR89 pKa = 11.84KK90 pKa = 9.17RR91 pKa = 11.84KK92 pKa = 9.36HH93 pKa = 5.58GFLARR98 pKa = 11.84LRR100 pKa = 11.84SAGGRR105 pKa = 11.84KK106 pKa = 8.73ILARR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84AKK114 pKa = 9.28GRR116 pKa = 11.84KK117 pKa = 8.14YY118 pKa = 10.54LSHH121 pKa = 7.04
MM1 pKa = 7.39PRR3 pKa = 11.84IPFSLSRR10 pKa = 11.84LLARR14 pKa = 11.84PLPIVQRR21 pKa = 11.84QPLVNRR27 pKa = 11.84SVSLTTHH34 pKa = 7.08LSSSIPRR41 pKa = 11.84LSSSPILRR49 pKa = 11.84AVSLLRR55 pKa = 11.84SSISLLPTTPTASHH69 pKa = 7.22LLQVRR74 pKa = 11.84FNQRR78 pKa = 11.84GVDD81 pKa = 3.75YY82 pKa = 10.92QPSQRR87 pKa = 11.84KK88 pKa = 8.81RR89 pKa = 11.84KK90 pKa = 9.17RR91 pKa = 11.84KK92 pKa = 9.36HH93 pKa = 5.58GFLARR98 pKa = 11.84LRR100 pKa = 11.84SAGGRR105 pKa = 11.84KK106 pKa = 8.73ILARR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84AKK114 pKa = 9.28GRR116 pKa = 11.84KK117 pKa = 8.14YY118 pKa = 10.54LSHH121 pKa = 7.04
Molecular weight: 13.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5547635 |
50 |
4925 |
425.5 |
47.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.036 ± 0.018 | 1.249 ± 0.009 |
5.567 ± 0.012 | 5.937 ± 0.018 |
3.873 ± 0.014 | 6.161 ± 0.021 |
2.605 ± 0.009 | 5.257 ± 0.015 |
4.507 ± 0.017 | 9.582 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.971 ± 0.007 | 3.483 ± 0.011 |
6.556 ± 0.029 | 3.687 ± 0.013 |
6.157 ± 0.017 | 9.156 ± 0.03 |
6.009 ± 0.014 | 6.123 ± 0.014 |
1.477 ± 0.008 | 2.606 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
