
Corynebacterium halotolerans YIM 70093 = DSM 44683
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Corynebacteriaceae; Corynebacterium; Corynebacterium halotolerans
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
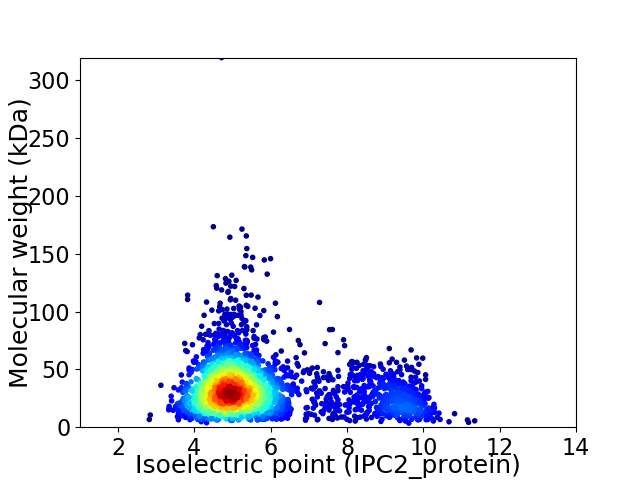
Virtual 2D-PAGE plot for 2853 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M1MYC0|M1MYC0_9CORY Membrane protein OS=Corynebacterium halotolerans YIM 70093 = DSM 44683 OX=1121362 GN=A605_08660 PE=4 SV=1
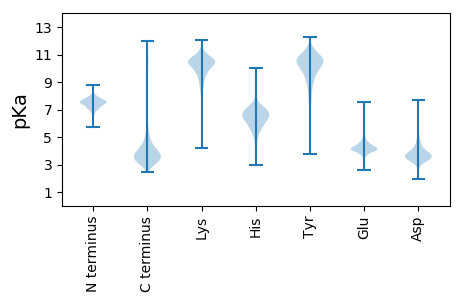
MM1 pKa = 7.8SGLTLAATGVLAPTVTAQEE20 pKa = 4.32DD21 pKa = 4.24TATTAWEE28 pKa = 3.88EE29 pKa = 3.64TAYY32 pKa = 10.37RR33 pKa = 11.84GSVEE37 pKa = 4.68VVDD40 pKa = 4.52EE41 pKa = 4.16QDD43 pKa = 5.11DD44 pKa = 4.26EE45 pKa = 4.49NTLEE49 pKa = 4.13GVVFEE54 pKa = 4.84DD55 pKa = 3.94TNKK58 pKa = 10.3NSQRR62 pKa = 11.84DD63 pKa = 3.62EE64 pKa = 4.42GEE66 pKa = 3.69PGIPGVSVSNGRR78 pKa = 11.84TIVTTDD84 pKa = 2.67AEE86 pKa = 4.49GRR88 pKa = 11.84YY89 pKa = 8.76EE90 pKa = 4.62LQVDD94 pKa = 3.88DD95 pKa = 4.28NTTVFITQPAGYY107 pKa = 9.0QVPVDD112 pKa = 4.28EE113 pKa = 5.13NNVAQFFYY121 pKa = 10.87NHH123 pKa = 5.73VPEE126 pKa = 5.05GSPDD130 pKa = 3.27LRR132 pKa = 11.84YY133 pKa = 10.55SGIEE137 pKa = 3.79PTGPLPDD144 pKa = 3.76AVNFPLSPSEE154 pKa = 4.02GTASPEE160 pKa = 3.94QNCVIGGDD168 pKa = 3.41IQTYY172 pKa = 5.57TAEE175 pKa = 4.11EE176 pKa = 3.92VEE178 pKa = 4.48YY179 pKa = 11.02ARR181 pKa = 11.84NGAFADD187 pKa = 4.12LAARR191 pKa = 11.84DD192 pKa = 4.47DD193 pKa = 4.05YY194 pKa = 11.8ANCGALFIGDD204 pKa = 3.83VVGDD208 pKa = 4.43DD209 pKa = 3.6LSLYY213 pKa = 10.08PDD215 pKa = 3.53VKK217 pKa = 10.75EE218 pKa = 3.91LTGMLNGPARR228 pKa = 11.84FLPGNHH234 pKa = 7.08DD235 pKa = 4.3LDD237 pKa = 4.62FDD239 pKa = 5.63ASDD242 pKa = 4.07EE243 pKa = 4.03DD244 pKa = 4.42HH245 pKa = 7.2KK246 pKa = 11.4FDD248 pKa = 3.75TYY250 pKa = 10.89RR251 pKa = 11.84SHH253 pKa = 7.73FGPEE257 pKa = 4.15YY258 pKa = 10.46YY259 pKa = 10.81SFDD262 pKa = 3.26VGNAHH267 pKa = 6.16VVALEE272 pKa = 4.03SVQYY276 pKa = 10.18PLEE279 pKa = 5.54DD280 pKa = 3.88GDD282 pKa = 4.76GYY284 pKa = 11.56NGAIDD289 pKa = 5.05DD290 pKa = 4.69EE291 pKa = 4.28QLEE294 pKa = 4.4WLRR297 pKa = 11.84QDD299 pKa = 3.23IAHH302 pKa = 6.12TPEE305 pKa = 4.88DD306 pKa = 4.05KK307 pKa = 11.18LIVLATHH314 pKa = 6.08VPLMSFADD322 pKa = 3.61QGSDD326 pKa = 2.99VHH328 pKa = 8.11QIDD331 pKa = 3.85QVQEE335 pKa = 3.61IYY337 pKa = 10.72EE338 pKa = 4.31IIGDD342 pKa = 3.93RR343 pKa = 11.84EE344 pKa = 4.21AIAVSGHH351 pKa = 3.93THH353 pKa = 6.69ALANMRR359 pKa = 11.84EE360 pKa = 4.07GDD362 pKa = 4.2SLEE365 pKa = 4.14GWNEE369 pKa = 3.84LYY371 pKa = 10.66GVEE374 pKa = 4.51EE375 pKa = 4.65LPFTHH380 pKa = 6.63IVAGAISGDD389 pKa = 3.6WYY391 pKa = 11.09SGEE394 pKa = 4.2TTEE397 pKa = 5.2NGYY400 pKa = 8.81PQAVGRR406 pKa = 11.84DD407 pKa = 3.51GSLPGVLTLDD417 pKa = 3.19IDD419 pKa = 3.93GSEE422 pKa = 3.87VSEE425 pKa = 4.96FYY427 pKa = 9.99TVRR430 pKa = 11.84GEE432 pKa = 4.19DD433 pKa = 3.37QSTQMALSLNTPAYY447 pKa = 9.91RR448 pKa = 11.84DD449 pKa = 3.47WYY451 pKa = 10.08EE452 pKa = 4.67DD453 pKa = 3.69NIDD456 pKa = 3.69NAGEE460 pKa = 4.07AEE462 pKa = 4.37EE463 pKa = 4.53FADD466 pKa = 3.67PLVVPEE472 pKa = 5.06DD473 pKa = 3.84EE474 pKa = 5.1VAEE477 pKa = 4.33TWLTTNFWMGSTGSTVEE494 pKa = 3.71VSIDD498 pKa = 3.18GGEE501 pKa = 3.8AVAAEE506 pKa = 4.67RR507 pKa = 11.84TQQMDD512 pKa = 3.76GEE514 pKa = 4.64VPNVGAEE521 pKa = 3.94WSDD524 pKa = 3.44PAAVQQQLVHH534 pKa = 6.47GGSVADD540 pKa = 4.32RR541 pKa = 11.84AAHH544 pKa = 6.53IWRR547 pKa = 11.84LPLPADD553 pKa = 4.37LGAGEE558 pKa = 4.37HH559 pKa = 5.13TAEE562 pKa = 4.07VTATDD567 pKa = 3.71VHH569 pKa = 6.12GHH571 pKa = 6.59EE572 pKa = 4.46YY573 pKa = 10.7TEE575 pKa = 4.27TLTFTVSPVEE585 pKa = 4.38GGDD588 pKa = 4.58DD589 pKa = 3.88NGDD592 pKa = 4.14DD593 pKa = 3.9DD594 pKa = 4.95TEE596 pKa = 4.91APSSGLSSLSSNLSSAGSLFF616 pKa = 3.54
MM1 pKa = 7.8SGLTLAATGVLAPTVTAQEE20 pKa = 4.32DD21 pKa = 4.24TATTAWEE28 pKa = 3.88EE29 pKa = 3.64TAYY32 pKa = 10.37RR33 pKa = 11.84GSVEE37 pKa = 4.68VVDD40 pKa = 4.52EE41 pKa = 4.16QDD43 pKa = 5.11DD44 pKa = 4.26EE45 pKa = 4.49NTLEE49 pKa = 4.13GVVFEE54 pKa = 4.84DD55 pKa = 3.94TNKK58 pKa = 10.3NSQRR62 pKa = 11.84DD63 pKa = 3.62EE64 pKa = 4.42GEE66 pKa = 3.69PGIPGVSVSNGRR78 pKa = 11.84TIVTTDD84 pKa = 2.67AEE86 pKa = 4.49GRR88 pKa = 11.84YY89 pKa = 8.76EE90 pKa = 4.62LQVDD94 pKa = 3.88DD95 pKa = 4.28NTTVFITQPAGYY107 pKa = 9.0QVPVDD112 pKa = 4.28EE113 pKa = 5.13NNVAQFFYY121 pKa = 10.87NHH123 pKa = 5.73VPEE126 pKa = 5.05GSPDD130 pKa = 3.27LRR132 pKa = 11.84YY133 pKa = 10.55SGIEE137 pKa = 3.79PTGPLPDD144 pKa = 3.76AVNFPLSPSEE154 pKa = 4.02GTASPEE160 pKa = 3.94QNCVIGGDD168 pKa = 3.41IQTYY172 pKa = 5.57TAEE175 pKa = 4.11EE176 pKa = 3.92VEE178 pKa = 4.48YY179 pKa = 11.02ARR181 pKa = 11.84NGAFADD187 pKa = 4.12LAARR191 pKa = 11.84DD192 pKa = 4.47DD193 pKa = 4.05YY194 pKa = 11.8ANCGALFIGDD204 pKa = 3.83VVGDD208 pKa = 4.43DD209 pKa = 3.6LSLYY213 pKa = 10.08PDD215 pKa = 3.53VKK217 pKa = 10.75EE218 pKa = 3.91LTGMLNGPARR228 pKa = 11.84FLPGNHH234 pKa = 7.08DD235 pKa = 4.3LDD237 pKa = 4.62FDD239 pKa = 5.63ASDD242 pKa = 4.07EE243 pKa = 4.03DD244 pKa = 4.42HH245 pKa = 7.2KK246 pKa = 11.4FDD248 pKa = 3.75TYY250 pKa = 10.89RR251 pKa = 11.84SHH253 pKa = 7.73FGPEE257 pKa = 4.15YY258 pKa = 10.46YY259 pKa = 10.81SFDD262 pKa = 3.26VGNAHH267 pKa = 6.16VVALEE272 pKa = 4.03SVQYY276 pKa = 10.18PLEE279 pKa = 5.54DD280 pKa = 3.88GDD282 pKa = 4.76GYY284 pKa = 11.56NGAIDD289 pKa = 5.05DD290 pKa = 4.69EE291 pKa = 4.28QLEE294 pKa = 4.4WLRR297 pKa = 11.84QDD299 pKa = 3.23IAHH302 pKa = 6.12TPEE305 pKa = 4.88DD306 pKa = 4.05KK307 pKa = 11.18LIVLATHH314 pKa = 6.08VPLMSFADD322 pKa = 3.61QGSDD326 pKa = 2.99VHH328 pKa = 8.11QIDD331 pKa = 3.85QVQEE335 pKa = 3.61IYY337 pKa = 10.72EE338 pKa = 4.31IIGDD342 pKa = 3.93RR343 pKa = 11.84EE344 pKa = 4.21AIAVSGHH351 pKa = 3.93THH353 pKa = 6.69ALANMRR359 pKa = 11.84EE360 pKa = 4.07GDD362 pKa = 4.2SLEE365 pKa = 4.14GWNEE369 pKa = 3.84LYY371 pKa = 10.66GVEE374 pKa = 4.51EE375 pKa = 4.65LPFTHH380 pKa = 6.63IVAGAISGDD389 pKa = 3.6WYY391 pKa = 11.09SGEE394 pKa = 4.2TTEE397 pKa = 5.2NGYY400 pKa = 8.81PQAVGRR406 pKa = 11.84DD407 pKa = 3.51GSLPGVLTLDD417 pKa = 3.19IDD419 pKa = 3.93GSEE422 pKa = 3.87VSEE425 pKa = 4.96FYY427 pKa = 9.99TVRR430 pKa = 11.84GEE432 pKa = 4.19DD433 pKa = 3.37QSTQMALSLNTPAYY447 pKa = 9.91RR448 pKa = 11.84DD449 pKa = 3.47WYY451 pKa = 10.08EE452 pKa = 4.67DD453 pKa = 3.69NIDD456 pKa = 3.69NAGEE460 pKa = 4.07AEE462 pKa = 4.37EE463 pKa = 4.53FADD466 pKa = 3.67PLVVPEE472 pKa = 5.06DD473 pKa = 3.84EE474 pKa = 5.1VAEE477 pKa = 4.33TWLTTNFWMGSTGSTVEE494 pKa = 3.71VSIDD498 pKa = 3.18GGEE501 pKa = 3.8AVAAEE506 pKa = 4.67RR507 pKa = 11.84TQQMDD512 pKa = 3.76GEE514 pKa = 4.64VPNVGAEE521 pKa = 3.94WSDD524 pKa = 3.44PAAVQQQLVHH534 pKa = 6.47GGSVADD540 pKa = 4.32RR541 pKa = 11.84AAHH544 pKa = 6.53IWRR547 pKa = 11.84LPLPADD553 pKa = 4.37LGAGEE558 pKa = 4.37HH559 pKa = 5.13TAEE562 pKa = 4.07VTATDD567 pKa = 3.71VHH569 pKa = 6.12GHH571 pKa = 6.59EE572 pKa = 4.46YY573 pKa = 10.7TEE575 pKa = 4.27TLTFTVSPVEE585 pKa = 4.38GGDD588 pKa = 4.58DD589 pKa = 3.88NGDD592 pKa = 4.14DD593 pKa = 3.9DD594 pKa = 4.95TEE596 pKa = 4.91APSSGLSSLSSNLSSAGSLFF616 pKa = 3.54
Molecular weight: 66.3 kDa
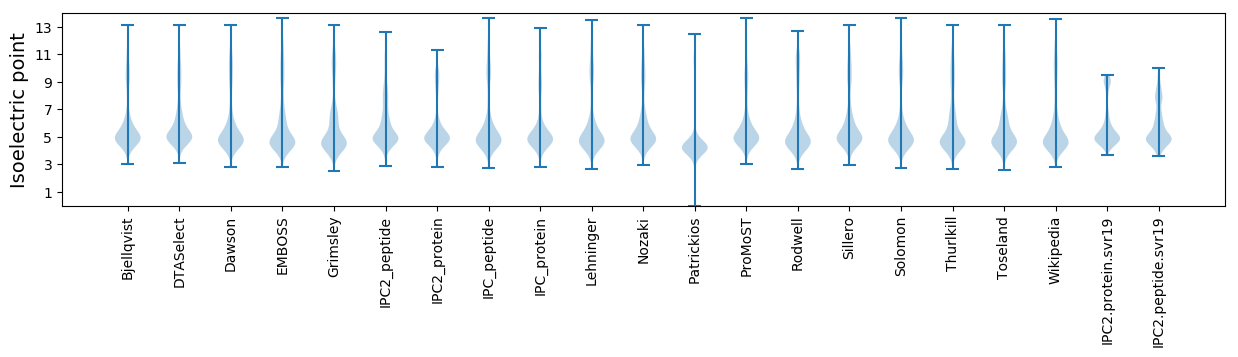
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M1N1M5|M1N1M5_9CORY Uncharacterized protein OS=Corynebacterium halotolerans YIM 70093 = DSM 44683 OX=1121362 GN=A605_14212 PE=4 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84TRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.73GRR42 pKa = 11.84AKK44 pKa = 9.63LTAA47 pKa = 4.21
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84TRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.73GRR42 pKa = 11.84AKK44 pKa = 9.63LTAA47 pKa = 4.21
Molecular weight: 5.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
924155 |
33 |
3023 |
323.9 |
34.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.941 ± 0.06 | 0.601 ± 0.011 |
6.302 ± 0.042 | 6.449 ± 0.048 |
3.137 ± 0.026 | 9.066 ± 0.044 |
2.259 ± 0.024 | 4.513 ± 0.034 |
2.235 ± 0.036 | 10.062 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.098 ± 0.02 | 2.371 ± 0.024 |
5.402 ± 0.036 | 2.901 ± 0.024 |
7.035 ± 0.043 | 5.242 ± 0.03 |
6.331 ± 0.032 | 8.576 ± 0.047 |
1.469 ± 0.021 | 2.01 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
