
Thiotrichales bacterium HS_08
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; unclassified Thiotrichales
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
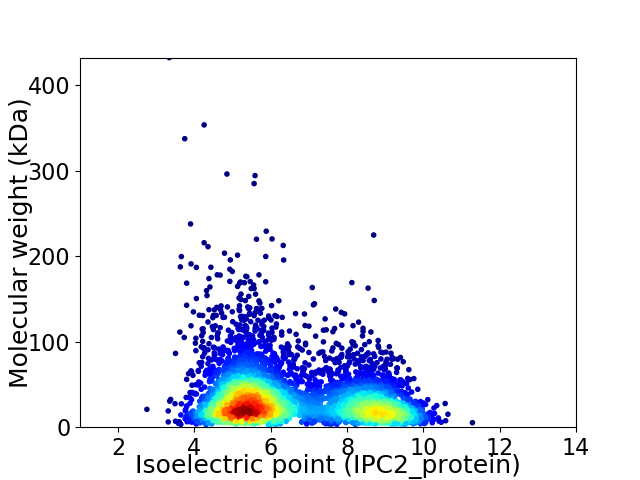
Virtual 2D-PAGE plot for 4918 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H6FC32|A0A1H6FC32_9GAMM Abequosyltransferase RfbV OS=Thiotrichales bacterium HS_08 OX=1899563 GN=rfbV PE=4 SV=1
MM1 pKa = 7.87KK2 pKa = 10.23IPITMRR8 pKa = 11.84QLLLFGSCMTLAGWMNTASADD29 pKa = 3.56VCVPTDD35 pKa = 3.66PNYY38 pKa = 11.14NPDD41 pKa = 5.16DD42 pKa = 5.16PICLTNPPPCDD53 pKa = 3.45PNVNPACQDD62 pKa = 3.31PVEE65 pKa = 5.03PPACDD70 pKa = 3.55PNVNPACQDD79 pKa = 3.31PVEE82 pKa = 5.03PPACDD87 pKa = 3.55PNVNPACQDD96 pKa = 3.31PVEE99 pKa = 5.03PPACDD104 pKa = 3.55PNVNPACQDD113 pKa = 3.31PVEE116 pKa = 5.03PPACDD121 pKa = 3.55PNVNPACQDD130 pKa = 3.26PVDD133 pKa = 4.55PQACDD138 pKa = 3.67PNVDD142 pKa = 4.16PACQDD147 pKa = 3.32SVDD150 pKa = 4.48PAACDD155 pKa = 3.59PNVNPGCTTNTPPPIDD171 pKa = 4.07CSLLINKK178 pKa = 8.57DD179 pKa = 3.33MPEE182 pKa = 4.24CTGEE186 pKa = 4.05QPKK189 pKa = 10.26FCMDD193 pKa = 3.14APEE196 pKa = 5.09RR197 pKa = 11.84GLFCDD202 pKa = 3.58EE203 pKa = 5.14FGHH206 pKa = 6.03KK207 pKa = 9.33VCRR210 pKa = 11.84VDD212 pKa = 5.11LIRR215 pKa = 11.84PGFLSCEE222 pKa = 4.18DD223 pKa = 3.56VPGVCVEE230 pKa = 4.77DD231 pKa = 3.78PSKK234 pKa = 11.4NLFCDD239 pKa = 3.3EE240 pKa = 4.95RR241 pKa = 11.84GKK243 pKa = 9.46QLCFPDD249 pKa = 3.87EE250 pKa = 4.19EE251 pKa = 5.18LGISCGDD258 pKa = 3.64VQCEE262 pKa = 3.97EE263 pKa = 4.45NPDD266 pKa = 3.25EE267 pKa = 6.34GIYY270 pKa = 10.58CDD272 pKa = 4.45AKK274 pKa = 10.47GHH276 pKa = 6.55PICEE280 pKa = 4.11EE281 pKa = 4.03NPLKK285 pKa = 10.9GIFCHH290 pKa = 6.91EE291 pKa = 4.32IPPACIHH298 pKa = 7.15DD299 pKa = 4.64PDD301 pKa = 5.03NGMTCDD307 pKa = 3.6ANHH310 pKa = 6.14NQGFVCIDD318 pKa = 3.84DD319 pKa = 4.38NDD321 pKa = 4.19PNNNVSCDD329 pKa = 3.62DD330 pKa = 3.56QGRR333 pKa = 11.84LCVTIPSGEE342 pKa = 4.37TFCEE346 pKa = 3.78EE347 pKa = 5.29DD348 pKa = 3.91GLFDD352 pKa = 4.01PAQFNSPDD360 pKa = 4.43LIVDD364 pKa = 3.92PALFNFFDD372 pKa = 4.54EE373 pKa = 4.5EE374 pKa = 5.91DD375 pKa = 3.55ITQLPPEE382 pKa = 4.42FFIGITPEE390 pKa = 3.74QFDD393 pKa = 3.94EE394 pKa = 4.65FRR396 pKa = 11.84PDD398 pKa = 3.57AYY400 pKa = 10.76GHH402 pKa = 6.68LNSGQVQALPLDD414 pKa = 3.66VLMDD418 pKa = 3.44MDD420 pKa = 3.85GQEE423 pKa = 4.75FFNFPPDD430 pKa = 3.39VFDD433 pKa = 6.53DD434 pKa = 4.91FSQEE438 pKa = 4.0QIGALPPKK446 pKa = 9.76ALRR449 pKa = 11.84GLTPTQFNRR458 pKa = 11.84LNPDD462 pKa = 3.06ALQGLDD468 pKa = 3.11RR469 pKa = 11.84HH470 pKa = 6.86DD471 pKa = 3.76IAALPPEE478 pKa = 4.59LWMDD482 pKa = 3.66IQQDD486 pKa = 4.09VFFGLPDD493 pKa = 3.89DD494 pKa = 5.14AFQGMDD500 pKa = 4.65HH501 pKa = 7.13EE502 pKa = 5.13DD503 pKa = 4.13CEE505 pKa = 4.35NLPPLMFGQMDD516 pKa = 3.83AAKK519 pKa = 9.98VGNLGGDD526 pKa = 3.96CLMGMDD532 pKa = 5.1SEE534 pKa = 5.52QFMQLPDD541 pKa = 3.39AALAGLGADD550 pKa = 4.43HH551 pKa = 7.01LHH553 pKa = 6.37GLPPRR558 pKa = 11.84VFNDD562 pKa = 3.18WTPEE566 pKa = 3.96RR567 pKa = 11.84FDD569 pKa = 4.59ALPDD573 pKa = 3.54AALTQLAQEE582 pKa = 4.85NPFEE586 pKa = 4.05MLEE589 pKa = 3.89ILANLDD595 pKa = 3.43TIAADD600 pKa = 3.98GVSADD605 pKa = 4.42DD606 pKa = 3.77LALVEE611 pKa = 5.23GFLPPGVSIDD621 pKa = 3.89PNTGEE626 pKa = 5.26LLFTGNEE633 pKa = 3.86FDD635 pKa = 4.12GQRR638 pKa = 11.84LPMPILEE645 pKa = 4.61FGADD649 pKa = 3.51VFGSDD654 pKa = 2.8IDD656 pKa = 4.5FRR658 pKa = 11.84LEE660 pKa = 3.65PHH662 pKa = 6.2NLAKK666 pKa = 10.86GFGLGGAGEE675 pKa = 4.19PALDD679 pKa = 3.59QMDD682 pKa = 3.74RR683 pKa = 11.84VMADD687 pKa = 3.42FFPGFGFKK695 pKa = 9.86QDD697 pKa = 3.36AEE699 pKa = 4.56GFISLDD705 pKa = 3.51GEE707 pKa = 4.42GDD709 pKa = 3.4FAGIGFDD716 pKa = 5.77LMLTPDD722 pKa = 5.47AIRR725 pKa = 11.84EE726 pKa = 4.31APEE729 pKa = 3.95GTPEE733 pKa = 5.52GITEE737 pKa = 4.2DD738 pKa = 3.65PMTGEE743 pKa = 4.17FLVTTDD749 pKa = 3.05HH750 pKa = 6.69GKK752 pKa = 9.75QFGVSFGPPNMTCMAKK768 pKa = 10.56ALGVGDD774 pKa = 4.0TNNPGSQLSMDD785 pKa = 3.72QNGIAVFMPATTSGTSNRR803 pKa = 11.84RR804 pKa = 11.84QFSNNPQSFKK814 pKa = 10.84PSGGSSNFRR823 pKa = 11.84RR824 pKa = 11.84SQQAAGLYY832 pKa = 7.23TYY834 pKa = 8.27PARR837 pKa = 11.84RR838 pKa = 11.84TGEE841 pKa = 3.87NPFAEE846 pKa = 4.36LVYY849 pKa = 10.77GQDD852 pKa = 3.27TGVEE856 pKa = 4.23EE857 pKa = 4.65CAGLTQTVYY866 pKa = 8.85PTVPDD871 pKa = 3.71ADD873 pKa = 4.36LFMEE877 pKa = 4.63MLANLLGIPTTDD889 pKa = 2.09IYY891 pKa = 11.46QRR893 pKa = 11.84VDD895 pKa = 2.8GSFRR899 pKa = 11.84VQLGPDD905 pKa = 3.76LYY907 pKa = 11.04DD908 pKa = 3.3IVPDD912 pKa = 4.56FVTEE916 pKa = 3.99APNIARR922 pKa = 11.84GTEE925 pKa = 3.89QATQLATDD933 pKa = 3.63AAGNFMMDD941 pKa = 2.97AQDD944 pKa = 4.12RR945 pKa = 11.84FILHH949 pKa = 7.12APYY952 pKa = 10.14QSRR955 pKa = 11.84KK956 pKa = 9.82LVIPLILKK964 pKa = 9.21VVVPQQ969 pKa = 3.66
MM1 pKa = 7.87KK2 pKa = 10.23IPITMRR8 pKa = 11.84QLLLFGSCMTLAGWMNTASADD29 pKa = 3.56VCVPTDD35 pKa = 3.66PNYY38 pKa = 11.14NPDD41 pKa = 5.16DD42 pKa = 5.16PICLTNPPPCDD53 pKa = 3.45PNVNPACQDD62 pKa = 3.31PVEE65 pKa = 5.03PPACDD70 pKa = 3.55PNVNPACQDD79 pKa = 3.31PVEE82 pKa = 5.03PPACDD87 pKa = 3.55PNVNPACQDD96 pKa = 3.31PVEE99 pKa = 5.03PPACDD104 pKa = 3.55PNVNPACQDD113 pKa = 3.31PVEE116 pKa = 5.03PPACDD121 pKa = 3.55PNVNPACQDD130 pKa = 3.26PVDD133 pKa = 4.55PQACDD138 pKa = 3.67PNVDD142 pKa = 4.16PACQDD147 pKa = 3.32SVDD150 pKa = 4.48PAACDD155 pKa = 3.59PNVNPGCTTNTPPPIDD171 pKa = 4.07CSLLINKK178 pKa = 8.57DD179 pKa = 3.33MPEE182 pKa = 4.24CTGEE186 pKa = 4.05QPKK189 pKa = 10.26FCMDD193 pKa = 3.14APEE196 pKa = 5.09RR197 pKa = 11.84GLFCDD202 pKa = 3.58EE203 pKa = 5.14FGHH206 pKa = 6.03KK207 pKa = 9.33VCRR210 pKa = 11.84VDD212 pKa = 5.11LIRR215 pKa = 11.84PGFLSCEE222 pKa = 4.18DD223 pKa = 3.56VPGVCVEE230 pKa = 4.77DD231 pKa = 3.78PSKK234 pKa = 11.4NLFCDD239 pKa = 3.3EE240 pKa = 4.95RR241 pKa = 11.84GKK243 pKa = 9.46QLCFPDD249 pKa = 3.87EE250 pKa = 4.19EE251 pKa = 5.18LGISCGDD258 pKa = 3.64VQCEE262 pKa = 3.97EE263 pKa = 4.45NPDD266 pKa = 3.25EE267 pKa = 6.34GIYY270 pKa = 10.58CDD272 pKa = 4.45AKK274 pKa = 10.47GHH276 pKa = 6.55PICEE280 pKa = 4.11EE281 pKa = 4.03NPLKK285 pKa = 10.9GIFCHH290 pKa = 6.91EE291 pKa = 4.32IPPACIHH298 pKa = 7.15DD299 pKa = 4.64PDD301 pKa = 5.03NGMTCDD307 pKa = 3.6ANHH310 pKa = 6.14NQGFVCIDD318 pKa = 3.84DD319 pKa = 4.38NDD321 pKa = 4.19PNNNVSCDD329 pKa = 3.62DD330 pKa = 3.56QGRR333 pKa = 11.84LCVTIPSGEE342 pKa = 4.37TFCEE346 pKa = 3.78EE347 pKa = 5.29DD348 pKa = 3.91GLFDD352 pKa = 4.01PAQFNSPDD360 pKa = 4.43LIVDD364 pKa = 3.92PALFNFFDD372 pKa = 4.54EE373 pKa = 4.5EE374 pKa = 5.91DD375 pKa = 3.55ITQLPPEE382 pKa = 4.42FFIGITPEE390 pKa = 3.74QFDD393 pKa = 3.94EE394 pKa = 4.65FRR396 pKa = 11.84PDD398 pKa = 3.57AYY400 pKa = 10.76GHH402 pKa = 6.68LNSGQVQALPLDD414 pKa = 3.66VLMDD418 pKa = 3.44MDD420 pKa = 3.85GQEE423 pKa = 4.75FFNFPPDD430 pKa = 3.39VFDD433 pKa = 6.53DD434 pKa = 4.91FSQEE438 pKa = 4.0QIGALPPKK446 pKa = 9.76ALRR449 pKa = 11.84GLTPTQFNRR458 pKa = 11.84LNPDD462 pKa = 3.06ALQGLDD468 pKa = 3.11RR469 pKa = 11.84HH470 pKa = 6.86DD471 pKa = 3.76IAALPPEE478 pKa = 4.59LWMDD482 pKa = 3.66IQQDD486 pKa = 4.09VFFGLPDD493 pKa = 3.89DD494 pKa = 5.14AFQGMDD500 pKa = 4.65HH501 pKa = 7.13EE502 pKa = 5.13DD503 pKa = 4.13CEE505 pKa = 4.35NLPPLMFGQMDD516 pKa = 3.83AAKK519 pKa = 9.98VGNLGGDD526 pKa = 3.96CLMGMDD532 pKa = 5.1SEE534 pKa = 5.52QFMQLPDD541 pKa = 3.39AALAGLGADD550 pKa = 4.43HH551 pKa = 7.01LHH553 pKa = 6.37GLPPRR558 pKa = 11.84VFNDD562 pKa = 3.18WTPEE566 pKa = 3.96RR567 pKa = 11.84FDD569 pKa = 4.59ALPDD573 pKa = 3.54AALTQLAQEE582 pKa = 4.85NPFEE586 pKa = 4.05MLEE589 pKa = 3.89ILANLDD595 pKa = 3.43TIAADD600 pKa = 3.98GVSADD605 pKa = 4.42DD606 pKa = 3.77LALVEE611 pKa = 5.23GFLPPGVSIDD621 pKa = 3.89PNTGEE626 pKa = 5.26LLFTGNEE633 pKa = 3.86FDD635 pKa = 4.12GQRR638 pKa = 11.84LPMPILEE645 pKa = 4.61FGADD649 pKa = 3.51VFGSDD654 pKa = 2.8IDD656 pKa = 4.5FRR658 pKa = 11.84LEE660 pKa = 3.65PHH662 pKa = 6.2NLAKK666 pKa = 10.86GFGLGGAGEE675 pKa = 4.19PALDD679 pKa = 3.59QMDD682 pKa = 3.74RR683 pKa = 11.84VMADD687 pKa = 3.42FFPGFGFKK695 pKa = 9.86QDD697 pKa = 3.36AEE699 pKa = 4.56GFISLDD705 pKa = 3.51GEE707 pKa = 4.42GDD709 pKa = 3.4FAGIGFDD716 pKa = 5.77LMLTPDD722 pKa = 5.47AIRR725 pKa = 11.84EE726 pKa = 4.31APEE729 pKa = 3.95GTPEE733 pKa = 5.52GITEE737 pKa = 4.2DD738 pKa = 3.65PMTGEE743 pKa = 4.17FLVTTDD749 pKa = 3.05HH750 pKa = 6.69GKK752 pKa = 9.75QFGVSFGPPNMTCMAKK768 pKa = 10.56ALGVGDD774 pKa = 4.0TNNPGSQLSMDD785 pKa = 3.72QNGIAVFMPATTSGTSNRR803 pKa = 11.84RR804 pKa = 11.84QFSNNPQSFKK814 pKa = 10.84PSGGSSNFRR823 pKa = 11.84RR824 pKa = 11.84SQQAAGLYY832 pKa = 7.23TYY834 pKa = 8.27PARR837 pKa = 11.84RR838 pKa = 11.84TGEE841 pKa = 3.87NPFAEE846 pKa = 4.36LVYY849 pKa = 10.77GQDD852 pKa = 3.27TGVEE856 pKa = 4.23EE857 pKa = 4.65CAGLTQTVYY866 pKa = 8.85PTVPDD871 pKa = 3.71ADD873 pKa = 4.36LFMEE877 pKa = 4.63MLANLLGIPTTDD889 pKa = 2.09IYY891 pKa = 11.46QRR893 pKa = 11.84VDD895 pKa = 2.8GSFRR899 pKa = 11.84VQLGPDD905 pKa = 3.76LYY907 pKa = 11.04DD908 pKa = 3.3IVPDD912 pKa = 4.56FVTEE916 pKa = 3.99APNIARR922 pKa = 11.84GTEE925 pKa = 3.89QATQLATDD933 pKa = 3.63AAGNFMMDD941 pKa = 2.97AQDD944 pKa = 4.12RR945 pKa = 11.84FILHH949 pKa = 7.12APYY952 pKa = 10.14QSRR955 pKa = 11.84KK956 pKa = 9.82LVIPLILKK964 pKa = 9.21VVVPQQ969 pKa = 3.66
Molecular weight: 104.9 kDa
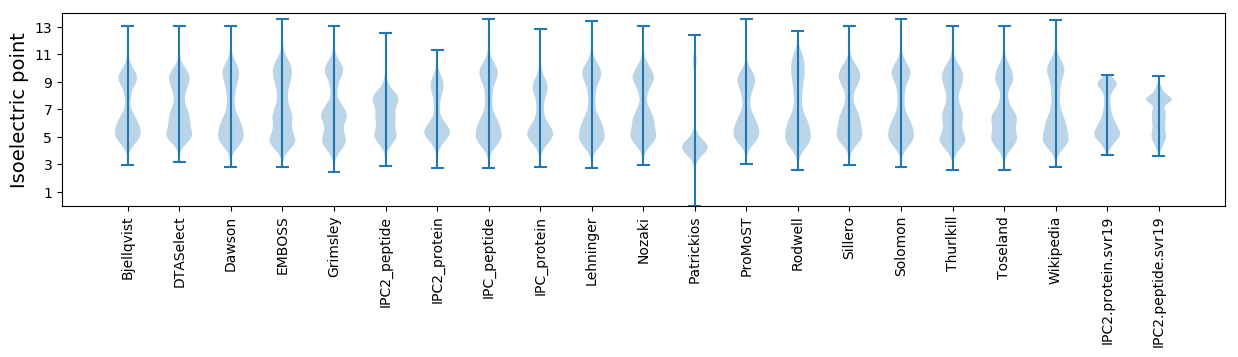
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6FCM3|A0A1H6FCM3_9GAMM Histidinol-phosphate aminotransferase OS=Thiotrichales bacterium HS_08 OX=1899563 GN=hisC2_1 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.54RR3 pKa = 11.84TFQPSQLVRR12 pKa = 11.84KK13 pKa = 9.03RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MQTRR25 pKa = 11.84GGRR28 pKa = 11.84QILRR32 pKa = 11.84NRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.45GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.54RR3 pKa = 11.84TFQPSQLVRR12 pKa = 11.84KK13 pKa = 9.03RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MQTRR25 pKa = 11.84GGRR28 pKa = 11.84QILRR32 pKa = 11.84NRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.45GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1586916 |
29 |
4142 |
322.7 |
36.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.354 ± 0.042 | 1.053 ± 0.013 |
5.419 ± 0.034 | 6.108 ± 0.04 |
4.04 ± 0.023 | 6.295 ± 0.043 |
2.514 ± 0.022 | 6.392 ± 0.032 |
5.463 ± 0.046 | 11.129 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.291 ± 0.017 | 4.47 ± 0.031 |
4.203 ± 0.024 | 5.634 ± 0.044 |
4.759 ± 0.034 | 6.153 ± 0.032 |
5.276 ± 0.039 | 5.775 ± 0.034 |
1.382 ± 0.016 | 3.254 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
