
Thioclava dalianensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Thioclava
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
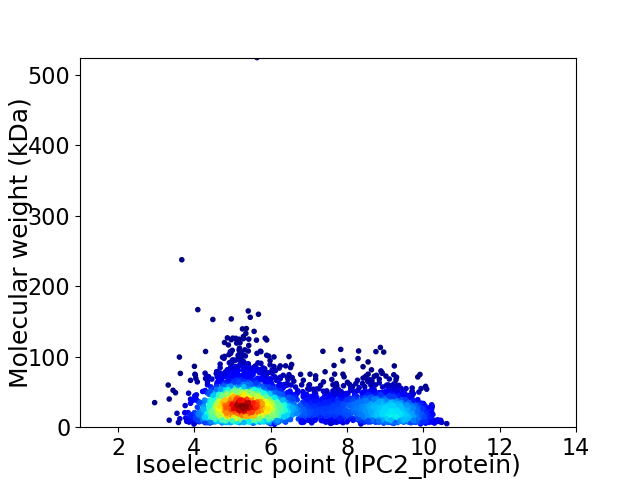
Virtual 2D-PAGE plot for 4007 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A074TA72|A0A074TA72_9RHOB Cytochrome c domain-containing protein OS=Thioclava dalianensis OX=1185766 GN=DL1_09710 PE=4 SV=1
MM1 pKa = 7.07TASPDD6 pKa = 3.62EE7 pKa = 4.44PLEE10 pKa = 4.12GAPLIKK16 pKa = 10.06PSSTDD21 pKa = 2.97HH22 pKa = 6.87PLYY25 pKa = 11.02DD26 pKa = 4.07PVVDD30 pKa = 4.07ACRR33 pKa = 11.84TVYY36 pKa = 10.75DD37 pKa = 3.83PEE39 pKa = 5.17IPVNIYY45 pKa = 10.8DD46 pKa = 4.42LGLIYY51 pKa = 10.15TIEE54 pKa = 3.88ITPEE58 pKa = 3.47NDD60 pKa = 2.89AKK62 pKa = 11.32VIMTLTAPGCPVAGEE77 pKa = 4.16MPGWVHH83 pKa = 7.19DD84 pKa = 4.33AVASVGGLRR93 pKa = 11.84DD94 pKa = 3.17VDD96 pKa = 3.83VEE98 pKa = 4.49MTFDD102 pKa = 4.09PQWGMDD108 pKa = 3.61MMSDD112 pKa = 3.68EE113 pKa = 4.89ARR115 pKa = 11.84LEE117 pKa = 4.14LGFMM121 pKa = 4.54
MM1 pKa = 7.07TASPDD6 pKa = 3.62EE7 pKa = 4.44PLEE10 pKa = 4.12GAPLIKK16 pKa = 10.06PSSTDD21 pKa = 2.97HH22 pKa = 6.87PLYY25 pKa = 11.02DD26 pKa = 4.07PVVDD30 pKa = 4.07ACRR33 pKa = 11.84TVYY36 pKa = 10.75DD37 pKa = 3.83PEE39 pKa = 5.17IPVNIYY45 pKa = 10.8DD46 pKa = 4.42LGLIYY51 pKa = 10.15TIEE54 pKa = 3.88ITPEE58 pKa = 3.47NDD60 pKa = 2.89AKK62 pKa = 11.32VIMTLTAPGCPVAGEE77 pKa = 4.16MPGWVHH83 pKa = 7.19DD84 pKa = 4.33AVASVGGLRR93 pKa = 11.84DD94 pKa = 3.17VDD96 pKa = 3.83VEE98 pKa = 4.49MTFDD102 pKa = 4.09PQWGMDD108 pKa = 3.61MMSDD112 pKa = 3.68EE113 pKa = 4.89ARR115 pKa = 11.84LEE117 pKa = 4.14LGFMM121 pKa = 4.54
Molecular weight: 13.17 kDa
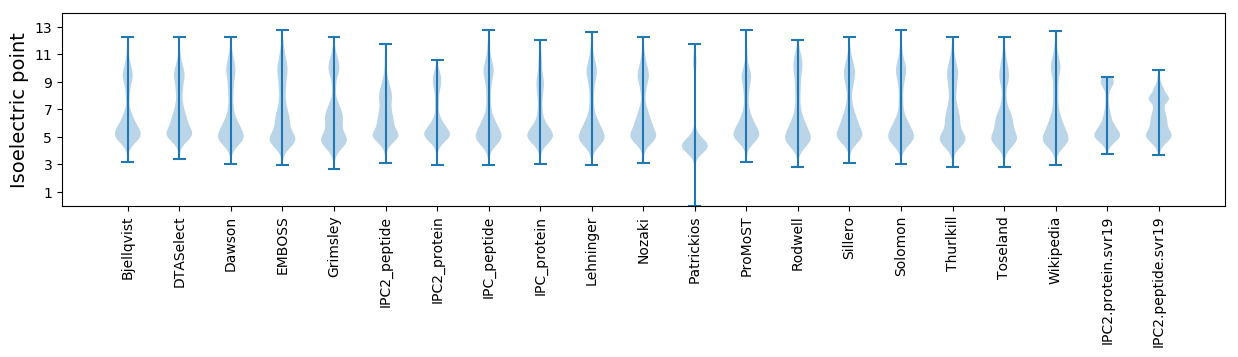
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A074TBQ2|A0A074TBQ2_9RHOB Signal peptidase I OS=Thioclava dalianensis OX=1185766 GN=DL1_05505 PE=3 SV=1
MM1 pKa = 7.54TNGPDD6 pKa = 3.25IASIAALIADD16 pKa = 4.55PARR19 pKa = 11.84ALMLNALMAGRR30 pKa = 11.84ALSAGEE36 pKa = 3.8LAGIAGIAPSTASSHH51 pKa = 6.25LARR54 pKa = 11.84LSAGGLVAMRR64 pKa = 11.84QQGRR68 pKa = 11.84HH69 pKa = 5.09RR70 pKa = 11.84YY71 pKa = 9.14FEE73 pKa = 4.2ISGSAAVEE81 pKa = 3.92LLEE84 pKa = 4.07SLMRR88 pKa = 11.84FSTVTLPRR96 pKa = 11.84LGPRR100 pKa = 11.84DD101 pKa = 3.28ADD103 pKa = 3.41LRR105 pKa = 11.84EE106 pKa = 3.93ARR108 pKa = 11.84LCYY111 pKa = 10.33NHH113 pKa = 6.98LAGRR117 pKa = 11.84AGVALYY123 pKa = 10.62DD124 pKa = 3.59GLIARR129 pKa = 11.84GALCPGHH136 pKa = 7.3DD137 pKa = 3.97GPEE140 pKa = 3.93LTEE143 pKa = 3.77TGRR146 pKa = 11.84AFVQTLGIDD155 pKa = 4.04LSAKK159 pKa = 6.85TFKK162 pKa = 10.63RR163 pKa = 11.84PPICRR168 pKa = 11.84SCLDD172 pKa = 2.99WSEE175 pKa = 4.31RR176 pKa = 11.84RR177 pKa = 11.84SHH179 pKa = 7.01LGGALGRR186 pKa = 11.84ALWEE190 pKa = 4.67AIATKK195 pKa = 10.08RR196 pKa = 11.84WARR199 pKa = 11.84QCGDD203 pKa = 3.0SRR205 pKa = 11.84VVRR208 pKa = 11.84FTPEE212 pKa = 3.41GQRR215 pKa = 11.84RR216 pKa = 11.84FAQLLAPKK224 pKa = 10.03TSMQGGKK231 pKa = 9.68SGQKK235 pKa = 9.57PRR237 pKa = 11.84LPAA240 pKa = 4.82
MM1 pKa = 7.54TNGPDD6 pKa = 3.25IASIAALIADD16 pKa = 4.55PARR19 pKa = 11.84ALMLNALMAGRR30 pKa = 11.84ALSAGEE36 pKa = 3.8LAGIAGIAPSTASSHH51 pKa = 6.25LARR54 pKa = 11.84LSAGGLVAMRR64 pKa = 11.84QQGRR68 pKa = 11.84HH69 pKa = 5.09RR70 pKa = 11.84YY71 pKa = 9.14FEE73 pKa = 4.2ISGSAAVEE81 pKa = 3.92LLEE84 pKa = 4.07SLMRR88 pKa = 11.84FSTVTLPRR96 pKa = 11.84LGPRR100 pKa = 11.84DD101 pKa = 3.28ADD103 pKa = 3.41LRR105 pKa = 11.84EE106 pKa = 3.93ARR108 pKa = 11.84LCYY111 pKa = 10.33NHH113 pKa = 6.98LAGRR117 pKa = 11.84AGVALYY123 pKa = 10.62DD124 pKa = 3.59GLIARR129 pKa = 11.84GALCPGHH136 pKa = 7.3DD137 pKa = 3.97GPEE140 pKa = 3.93LTEE143 pKa = 3.77TGRR146 pKa = 11.84AFVQTLGIDD155 pKa = 4.04LSAKK159 pKa = 6.85TFKK162 pKa = 10.63RR163 pKa = 11.84PPICRR168 pKa = 11.84SCLDD172 pKa = 2.99WSEE175 pKa = 4.31RR176 pKa = 11.84RR177 pKa = 11.84SHH179 pKa = 7.01LGGALGRR186 pKa = 11.84ALWEE190 pKa = 4.67AIATKK195 pKa = 10.08RR196 pKa = 11.84WARR199 pKa = 11.84QCGDD203 pKa = 3.0SRR205 pKa = 11.84VVRR208 pKa = 11.84FTPEE212 pKa = 3.41GQRR215 pKa = 11.84RR216 pKa = 11.84FAQLLAPKK224 pKa = 10.03TSMQGGKK231 pKa = 9.68SGQKK235 pKa = 9.57PRR237 pKa = 11.84LPAA240 pKa = 4.82
Molecular weight: 25.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1230192 |
39 |
4774 |
307.0 |
33.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.642 ± 0.052 | 0.869 ± 0.013 |
5.805 ± 0.033 | 5.8 ± 0.04 |
3.642 ± 0.023 | 8.64 ± 0.042 |
2.022 ± 0.019 | 5.241 ± 0.027 |
3.293 ± 0.035 | 10.188 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.763 ± 0.019 | 2.479 ± 0.022 |
5.23 ± 0.028 | 3.369 ± 0.024 |
6.794 ± 0.039 | 5.385 ± 0.028 |
5.312 ± 0.03 | 6.927 ± 0.035 |
1.418 ± 0.018 | 2.181 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
