
Hyalangium minutum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Cystobacterineae; Archangiaceae; Hyalangium
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
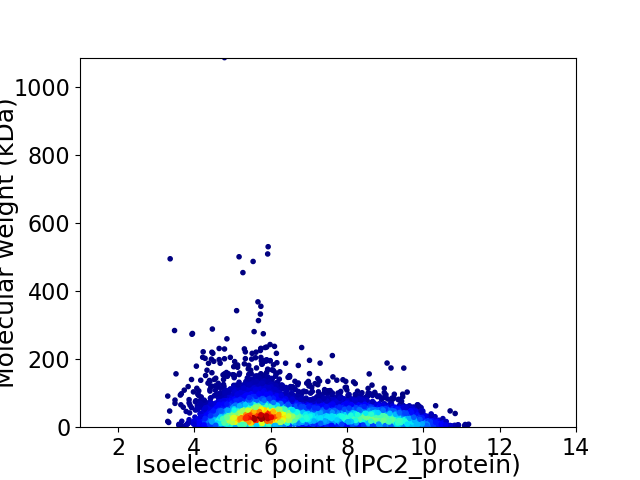
Virtual 2D-PAGE plot for 8967 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A085VZS1|A0A085VZS1_9DELT Chromosome partition protein smc OS=Hyalangium minutum OX=394096 GN=DB31_4558 PE=4 SV=1
MM1 pKa = 7.52RR2 pKa = 11.84RR3 pKa = 11.84SSSSSRR9 pKa = 11.84ISAVCSSLLAALGLLLSAPASAQWTPGTALPHH41 pKa = 6.18AHH43 pKa = 5.55TQGVIIPDD51 pKa = 3.28ATSRR55 pKa = 11.84IHH57 pKa = 7.3AIGGGDD63 pKa = 3.39SGAFDD68 pKa = 3.99TNAHH72 pKa = 6.76DD73 pKa = 3.97EE74 pKa = 4.4FDD76 pKa = 4.43PVTHH80 pKa = 5.98TWITRR85 pKa = 11.84APLPINNRR93 pKa = 11.84GMGASLGPDD102 pKa = 2.96GRR104 pKa = 11.84IYY106 pKa = 10.88VYY108 pKa = 10.9GGFAGWGSIGAVYY121 pKa = 10.03AYY123 pKa = 8.93TPSTDD128 pKa = 2.46TWAPLASMPTPQWEE142 pKa = 4.23TRR144 pKa = 11.84GDD146 pKa = 3.52FGSDD150 pKa = 2.39GKK152 pKa = 11.28LYY154 pKa = 11.03VFGGEE159 pKa = 4.54TNGSASAVTQIYY171 pKa = 8.34TPSTNTWTTGASMPGGRR188 pKa = 11.84RR189 pKa = 11.84QHH191 pKa = 5.56GAFRR195 pKa = 11.84GSDD198 pKa = 2.83GRR200 pKa = 11.84FYY202 pKa = 11.57VVGGQDD208 pKa = 3.03ASAATAGTYY217 pKa = 9.6IYY219 pKa = 11.06NPTTNTWTTGPAMPAAANSFGQAVSTDD246 pKa = 3.04RR247 pKa = 11.84SRR249 pKa = 11.84FFVLGGSTSYY259 pKa = 11.47NNEE262 pKa = 3.83STPLFDD268 pKa = 3.15TVYY271 pKa = 9.07MFNVNTQEE279 pKa = 4.04WTSLTGFPTARR290 pKa = 11.84RR291 pKa = 11.84EE292 pKa = 3.88LGATVTHH299 pKa = 6.38CRR301 pKa = 11.84LRR303 pKa = 11.84TLGGSTGTSVTTHH316 pKa = 5.7EE317 pKa = 4.44VFTLTGAPDD326 pKa = 3.74TNGNGQPDD334 pKa = 4.14YY335 pKa = 11.59CEE337 pKa = 5.71DD338 pKa = 4.57PDD340 pKa = 4.96IDD342 pKa = 5.35DD343 pKa = 5.72DD344 pKa = 4.53NVPNSADD351 pKa = 3.43NCPTVPNPGQEE362 pKa = 4.28NQDD365 pKa = 3.19HH366 pKa = 7.17DD367 pKa = 4.43AQGDD371 pKa = 3.81ACDD374 pKa = 4.35PDD376 pKa = 4.72ADD378 pKa = 4.93NDD380 pKa = 4.18ALPNGQDD387 pKa = 3.29NCPLHH392 pKa = 6.88PNADD396 pKa = 3.84QTDD399 pKa = 3.57SDD401 pKa = 4.76ADD403 pKa = 3.9GLGNACDD410 pKa = 4.4PDD412 pKa = 3.74NDD414 pKa = 3.99NDD416 pKa = 3.93GTADD420 pKa = 4.44LSDD423 pKa = 3.81NCPSQSNADD432 pKa = 3.52QHH434 pKa = 6.67DD435 pKa = 3.81TDD437 pKa = 4.7GDD439 pKa = 4.53SIGDD443 pKa = 3.72ACDD446 pKa = 3.9SDD448 pKa = 5.21DD449 pKa = 5.72DD450 pKa = 4.61NDD452 pKa = 5.47SIADD456 pKa = 3.84PQDD459 pKa = 3.39NCPLVANTDD468 pKa = 3.38QRR470 pKa = 11.84DD471 pKa = 3.48TDD473 pKa = 3.73GDD475 pKa = 4.53SIGDD479 pKa = 3.76ACDD482 pKa = 3.6SDD484 pKa = 3.93TDD486 pKa = 4.01SDD488 pKa = 4.09NDD490 pKa = 4.02TVPDD494 pKa = 3.72VTDD497 pKa = 3.32NCALVPNPDD506 pKa = 3.07QANADD511 pKa = 3.67KK512 pKa = 11.14DD513 pKa = 4.17EE514 pKa = 4.68QGDD517 pKa = 3.87ACDD520 pKa = 5.24SDD522 pKa = 4.83DD523 pKa = 6.31DD524 pKa = 4.62NDD526 pKa = 4.03TVADD530 pKa = 3.89EE531 pKa = 4.35WDD533 pKa = 3.63NCPFIPNTDD542 pKa = 3.43QADD545 pKa = 3.43SDD547 pKa = 4.52GNGKK551 pKa = 10.56GDD553 pKa = 3.53VCDD556 pKa = 4.21VDD558 pKa = 4.7NDD560 pKa = 3.73TDD562 pKa = 4.15KK563 pKa = 11.81DD564 pKa = 3.92GVANATDD571 pKa = 3.73NCPLVANPDD580 pKa = 3.71QLDD583 pKa = 3.52SDD585 pKa = 4.24GDD587 pKa = 4.4GVGDD591 pKa = 4.02ACDD594 pKa = 3.6EE595 pKa = 4.49SGGGAGGCGGGCGAGPIGSSSLMLWALAGLALRR628 pKa = 11.84ARR630 pKa = 11.84RR631 pKa = 11.84RR632 pKa = 11.84FARR635 pKa = 3.78
MM1 pKa = 7.52RR2 pKa = 11.84RR3 pKa = 11.84SSSSSRR9 pKa = 11.84ISAVCSSLLAALGLLLSAPASAQWTPGTALPHH41 pKa = 6.18AHH43 pKa = 5.55TQGVIIPDD51 pKa = 3.28ATSRR55 pKa = 11.84IHH57 pKa = 7.3AIGGGDD63 pKa = 3.39SGAFDD68 pKa = 3.99TNAHH72 pKa = 6.76DD73 pKa = 3.97EE74 pKa = 4.4FDD76 pKa = 4.43PVTHH80 pKa = 5.98TWITRR85 pKa = 11.84APLPINNRR93 pKa = 11.84GMGASLGPDD102 pKa = 2.96GRR104 pKa = 11.84IYY106 pKa = 10.88VYY108 pKa = 10.9GGFAGWGSIGAVYY121 pKa = 10.03AYY123 pKa = 8.93TPSTDD128 pKa = 2.46TWAPLASMPTPQWEE142 pKa = 4.23TRR144 pKa = 11.84GDD146 pKa = 3.52FGSDD150 pKa = 2.39GKK152 pKa = 11.28LYY154 pKa = 11.03VFGGEE159 pKa = 4.54TNGSASAVTQIYY171 pKa = 8.34TPSTNTWTTGASMPGGRR188 pKa = 11.84RR189 pKa = 11.84QHH191 pKa = 5.56GAFRR195 pKa = 11.84GSDD198 pKa = 2.83GRR200 pKa = 11.84FYY202 pKa = 11.57VVGGQDD208 pKa = 3.03ASAATAGTYY217 pKa = 9.6IYY219 pKa = 11.06NPTTNTWTTGPAMPAAANSFGQAVSTDD246 pKa = 3.04RR247 pKa = 11.84SRR249 pKa = 11.84FFVLGGSTSYY259 pKa = 11.47NNEE262 pKa = 3.83STPLFDD268 pKa = 3.15TVYY271 pKa = 9.07MFNVNTQEE279 pKa = 4.04WTSLTGFPTARR290 pKa = 11.84RR291 pKa = 11.84EE292 pKa = 3.88LGATVTHH299 pKa = 6.38CRR301 pKa = 11.84LRR303 pKa = 11.84TLGGSTGTSVTTHH316 pKa = 5.7EE317 pKa = 4.44VFTLTGAPDD326 pKa = 3.74TNGNGQPDD334 pKa = 4.14YY335 pKa = 11.59CEE337 pKa = 5.71DD338 pKa = 4.57PDD340 pKa = 4.96IDD342 pKa = 5.35DD343 pKa = 5.72DD344 pKa = 4.53NVPNSADD351 pKa = 3.43NCPTVPNPGQEE362 pKa = 4.28NQDD365 pKa = 3.19HH366 pKa = 7.17DD367 pKa = 4.43AQGDD371 pKa = 3.81ACDD374 pKa = 4.35PDD376 pKa = 4.72ADD378 pKa = 4.93NDD380 pKa = 4.18ALPNGQDD387 pKa = 3.29NCPLHH392 pKa = 6.88PNADD396 pKa = 3.84QTDD399 pKa = 3.57SDD401 pKa = 4.76ADD403 pKa = 3.9GLGNACDD410 pKa = 4.4PDD412 pKa = 3.74NDD414 pKa = 3.99NDD416 pKa = 3.93GTADD420 pKa = 4.44LSDD423 pKa = 3.81NCPSQSNADD432 pKa = 3.52QHH434 pKa = 6.67DD435 pKa = 3.81TDD437 pKa = 4.7GDD439 pKa = 4.53SIGDD443 pKa = 3.72ACDD446 pKa = 3.9SDD448 pKa = 5.21DD449 pKa = 5.72DD450 pKa = 4.61NDD452 pKa = 5.47SIADD456 pKa = 3.84PQDD459 pKa = 3.39NCPLVANTDD468 pKa = 3.38QRR470 pKa = 11.84DD471 pKa = 3.48TDD473 pKa = 3.73GDD475 pKa = 4.53SIGDD479 pKa = 3.76ACDD482 pKa = 3.6SDD484 pKa = 3.93TDD486 pKa = 4.01SDD488 pKa = 4.09NDD490 pKa = 4.02TVPDD494 pKa = 3.72VTDD497 pKa = 3.32NCALVPNPDD506 pKa = 3.07QANADD511 pKa = 3.67KK512 pKa = 11.14DD513 pKa = 4.17EE514 pKa = 4.68QGDD517 pKa = 3.87ACDD520 pKa = 5.24SDD522 pKa = 4.83DD523 pKa = 6.31DD524 pKa = 4.62NDD526 pKa = 4.03TVADD530 pKa = 3.89EE531 pKa = 4.35WDD533 pKa = 3.63NCPFIPNTDD542 pKa = 3.43QADD545 pKa = 3.43SDD547 pKa = 4.52GNGKK551 pKa = 10.56GDD553 pKa = 3.53VCDD556 pKa = 4.21VDD558 pKa = 4.7NDD560 pKa = 3.73TDD562 pKa = 4.15KK563 pKa = 11.81DD564 pKa = 3.92GVANATDD571 pKa = 3.73NCPLVANPDD580 pKa = 3.71QLDD583 pKa = 3.52SDD585 pKa = 4.24GDD587 pKa = 4.4GVGDD591 pKa = 4.02ACDD594 pKa = 3.6EE595 pKa = 4.49SGGGAGGCGGGCGAGPIGSSSLMLWALAGLALRR628 pKa = 11.84ARR630 pKa = 11.84RR631 pKa = 11.84RR632 pKa = 11.84FARR635 pKa = 3.78
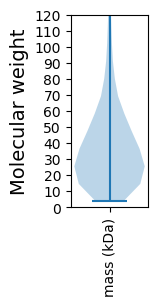
Molecular weight: 65.83 kDa
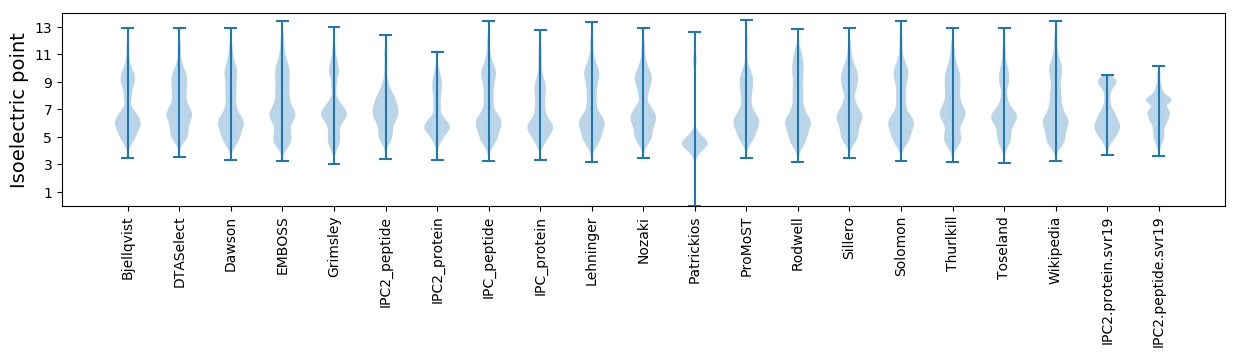
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A085WNG5|A0A085WNG5_9DELT Uncharacterized protein OS=Hyalangium minutum OX=394096 GN=DB31_7130 PE=4 SV=1
MM1 pKa = 7.07VRR3 pKa = 11.84GHH5 pKa = 5.61STSGFHH11 pKa = 7.19GNRR14 pKa = 11.84PSSRR18 pKa = 11.84FWPARR23 pKa = 11.84GALGQALHH31 pKa = 6.76GSAAAAPAQGAAGRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 6.16LSLSALRR54 pKa = 11.84SVLNAHH60 pKa = 7.28LSPLSSRR67 pKa = 11.84SSPSMAPARR76 pKa = 11.84PQSPPAHH83 pKa = 6.34RR84 pKa = 4.59
MM1 pKa = 7.07VRR3 pKa = 11.84GHH5 pKa = 5.61STSGFHH11 pKa = 7.19GNRR14 pKa = 11.84PSSRR18 pKa = 11.84FWPARR23 pKa = 11.84GALGQALHH31 pKa = 6.76GSAAAAPAQGAAGRR45 pKa = 11.84RR46 pKa = 11.84HH47 pKa = 6.16LSLSALRR54 pKa = 11.84SVLNAHH60 pKa = 7.28LSPLSSRR67 pKa = 11.84SSPSMAPARR76 pKa = 11.84PQSPPAHH83 pKa = 6.34RR84 pKa = 4.59
Molecular weight: 8.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3319859 |
37 |
10488 |
370.2 |
40.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.534 ± 0.037 | 0.944 ± 0.012 |
4.794 ± 0.02 | 6.591 ± 0.036 |
3.387 ± 0.018 | 8.568 ± 0.032 |
2.043 ± 0.012 | 3.239 ± 0.017 |
3.263 ± 0.027 | 10.931 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.946 ± 0.012 | 2.333 ± 0.021 |
6.215 ± 0.027 | 3.772 ± 0.016 |
7.472 ± 0.034 | 6.055 ± 0.027 |
5.592 ± 0.039 | 7.778 ± 0.028 |
1.358 ± 0.012 | 2.188 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
