
uncultured phage_MedDCM-OCT-S38-C3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Stopalavirus; Stopalavirus S38C3
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
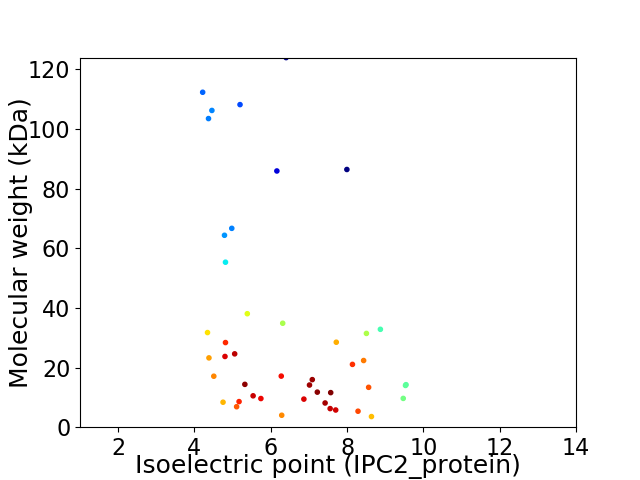
Virtual 2D-PAGE plot for 45 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A6S4PGX3|A0A6S4PGX3_9CAUD Uncharacterized protein OS=uncultured phage_MedDCM-OCT-S38-C3 OX=2740803 PE=4 SV=1
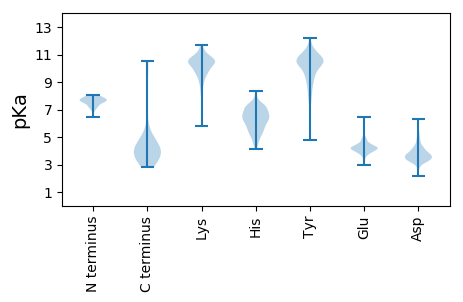
MM1 pKa = 7.49PRR3 pKa = 11.84QYY5 pKa = 11.43DD6 pKa = 3.87SNDD9 pKa = 3.18VNPFEE14 pKa = 5.81DD15 pKa = 4.38LGPLQFPPTDD25 pKa = 3.68EE26 pKa = 4.71NSPDD30 pKa = 4.23DD31 pKa = 3.89GSLIYY36 pKa = 10.47IHH38 pKa = 7.24GNTTYY43 pKa = 10.25IWDD46 pKa = 3.95GEE48 pKa = 4.21KK49 pKa = 8.34WTASTSGPRR58 pKa = 11.84LVDD61 pKa = 3.59LTWDD65 pKa = 3.93PNSGGGWLLNNAGADD80 pKa = 3.32VWFASVNEE88 pKa = 3.94VDD90 pKa = 4.48AGLMTPDD97 pKa = 2.68ILGRR101 pKa = 11.84IEE103 pKa = 4.48ALEE106 pKa = 4.02LHH108 pKa = 6.75GGAQGIVIKK117 pKa = 10.83GRR119 pKa = 11.84LDD121 pKa = 3.43SVGPPDD127 pKa = 4.1PSLAISVGDD136 pKa = 4.53AYY138 pKa = 11.44LDD140 pKa = 3.75TNGDD144 pKa = 3.24LWVCTEE150 pKa = 3.79EE151 pKa = 5.18LEE153 pKa = 4.35FVNVGMVQGPPGPQGPAGPQGPRR176 pKa = 11.84GYY178 pKa = 10.63DD179 pKa = 3.31GEE181 pKa = 4.68NGLDD185 pKa = 3.76GEE187 pKa = 4.71PGRR190 pKa = 11.84DD191 pKa = 3.65GTPGAQGPPGAPGRR205 pKa = 11.84DD206 pKa = 3.8GQPGAQGEE214 pKa = 4.12QGPQGEE220 pKa = 4.31KK221 pKa = 10.97GEE223 pKa = 4.49RR224 pKa = 11.84GWEE227 pKa = 4.28GATGPQGEE235 pKa = 4.7EE236 pKa = 4.79GPVGPQGPKK245 pKa = 10.04GDD247 pKa = 4.03RR248 pKa = 11.84GDD250 pKa = 3.79VGPEE254 pKa = 4.01GPVVGVEE261 pKa = 4.3GPEE264 pKa = 4.45GPVGPPGPEE273 pKa = 4.11GPEE276 pKa = 4.47GPDD279 pKa = 3.74GPQGPPGTSFANLKK293 pKa = 10.3GVVATEE299 pKa = 4.14HH300 pKa = 7.27DD301 pKa = 4.47LPADD305 pKa = 3.51PTQWDD310 pKa = 3.96MYY312 pKa = 10.59LVEE315 pKa = 4.98SADD318 pKa = 5.39KK319 pKa = 8.94YY320 pKa = 8.95TIWDD324 pKa = 3.77GEE326 pKa = 3.9KK327 pKa = 9.21WRR329 pKa = 11.84YY330 pKa = 7.89VAAGGSVEE338 pKa = 4.82DD339 pKa = 4.74APGDD343 pKa = 3.65SKK345 pKa = 11.5PYY347 pKa = 10.27VRR349 pKa = 11.84YY350 pKa = 10.7NMDD353 pKa = 2.47WRR355 pKa = 11.84EE356 pKa = 4.03GVLDD360 pKa = 4.72APDD363 pKa = 4.33DD364 pKa = 4.0GKK366 pKa = 11.65GYY368 pKa = 10.72VRR370 pKa = 11.84YY371 pKa = 8.28QQGWASLEE379 pKa = 4.21AVGLPYY385 pKa = 10.84LIGTDD390 pKa = 3.22KK391 pKa = 11.03TGLARR396 pKa = 11.84SVRR399 pKa = 11.84KK400 pKa = 9.89AGPFTNTKK408 pKa = 11.0DD409 pKa = 3.32MGPEE413 pKa = 3.45IEE415 pKa = 5.82LIDD418 pKa = 4.46GNGNYY423 pKa = 10.45SNVKK427 pKa = 9.15IGGSGGVVTSSDD439 pKa = 3.24AAGITVDD446 pKa = 4.86GSYY449 pKa = 11.12LLKK452 pKa = 10.7KK453 pKa = 10.57QFDD456 pKa = 4.35EE457 pKa = 4.71YY458 pKa = 10.24PTIDD462 pKa = 4.56AEE464 pKa = 4.53TLDD467 pKa = 4.3SVPADD472 pKa = 2.98SVMFAANQQGNKK484 pKa = 9.93SITWKK489 pKa = 10.8EE490 pKa = 3.7MMGSVMHH497 pKa = 6.24QIGFAIQSASITEE510 pKa = 4.32DD511 pKa = 3.22NAPAQIDD518 pKa = 3.63RR519 pKa = 11.84AVEE522 pKa = 4.36CTCQATLEE530 pKa = 4.27SSAPQFGEE538 pKa = 4.43DD539 pKa = 3.06VCTYY543 pKa = 8.88RR544 pKa = 11.84WRR546 pKa = 11.84AFPQGTSDD554 pKa = 3.32NAKK557 pKa = 10.29PEE559 pKa = 4.09WEE561 pKa = 4.08NGKK564 pKa = 9.66VPSNPNKK571 pKa = 10.05KK572 pKa = 10.41SFVFDD577 pKa = 3.29EE578 pKa = 4.82AGDD581 pKa = 3.77YY582 pKa = 10.57TIEE585 pKa = 4.54CEE587 pKa = 4.3VGASKK592 pKa = 10.89SKK594 pKa = 10.31LWNQVTKK601 pKa = 10.08VASYY605 pKa = 10.91DD606 pKa = 3.44FHH608 pKa = 8.3VKK610 pKa = 10.32EE611 pKa = 4.9GPDD614 pKa = 3.4PLMSYY619 pKa = 10.87AYY621 pKa = 10.32LHH623 pKa = 6.22VKK625 pKa = 10.26NITGGQIKK633 pKa = 9.8FPNALDD639 pKa = 4.1SGNHH643 pKa = 5.37YY644 pKa = 10.72YY645 pKa = 10.69DD646 pKa = 3.76WEE648 pKa = 4.46NMGHH652 pKa = 5.37EE653 pKa = 4.21PEE655 pKa = 4.88YY656 pKa = 10.36YY657 pKa = 10.77AFVHH661 pKa = 5.8VLGLEE666 pKa = 4.53GGDD669 pKa = 3.34QAFGGFKK676 pKa = 10.35CYY678 pKa = 10.56RR679 pKa = 11.84NVQGDD684 pKa = 3.35WDD686 pKa = 3.73DD687 pKa = 3.8WEE689 pKa = 4.46TDD691 pKa = 3.61KK692 pKa = 11.71SDD694 pKa = 3.68EE695 pKa = 4.24TCAKK699 pKa = 10.25YY700 pKa = 10.46GISFYY705 pKa = 11.52DD706 pKa = 3.18NDD708 pKa = 5.1LYY710 pKa = 11.47LEE712 pKa = 4.89PDD714 pKa = 3.0TGLEE718 pKa = 3.86YY719 pKa = 9.88MVYY722 pKa = 9.77MRR724 pKa = 11.84TQTYY728 pKa = 8.81NQVGGSDD735 pKa = 3.66DD736 pKa = 4.01SGVDD740 pKa = 3.54GLNQAGAWLDD750 pKa = 3.52GQRR753 pKa = 11.84WSTTGNATWEE763 pKa = 4.13WGEE766 pKa = 3.92LTDD769 pKa = 3.85TSKK772 pKa = 9.42VTSFYY777 pKa = 11.31RR778 pKa = 11.84MFEE781 pKa = 3.95QFSNSGSAWPTGWEE795 pKa = 4.12HH796 pKa = 7.06FDD798 pKa = 3.2TGKK801 pKa = 11.11GEE803 pKa = 3.8NFAWMFYY810 pKa = 7.6QARR813 pKa = 11.84ATGGIGCGMEE823 pKa = 3.86QWNMSKK829 pKa = 10.31ARR831 pKa = 11.84SLHH834 pKa = 5.48RR835 pKa = 11.84MFYY838 pKa = 9.38QAEE841 pKa = 4.14NFDD844 pKa = 4.53CDD846 pKa = 3.09ISNWDD851 pKa = 3.28TRR853 pKa = 11.84EE854 pKa = 3.84NEE856 pKa = 3.79TLYY859 pKa = 11.19YY860 pKa = 11.0FMGQCKK866 pKa = 10.02NFTCGGLNASEE877 pKa = 4.31EE878 pKa = 5.01GYY880 pKa = 8.66GTGIVNMKK888 pKa = 6.96TLKK891 pKa = 10.54VKK893 pKa = 10.19EE894 pKa = 4.2INYY897 pKa = 9.94SFYY900 pKa = 10.81QCEE903 pKa = 4.26KK904 pKa = 10.93LYY906 pKa = 10.31IDD908 pKa = 4.27CSQWGFPNLEE918 pKa = 4.0NFDD921 pKa = 4.27WDD923 pKa = 3.17TDD925 pKa = 3.73AAPDD929 pKa = 3.5VEE931 pKa = 4.86MFQTANIPRR940 pKa = 11.84SKK942 pKa = 11.01ASDD945 pKa = 3.26YY946 pKa = 10.92WGITSYY952 pKa = 11.52DD953 pKa = 3.84SYY955 pKa = 11.29WNVQIAGILPIWGTDD970 pKa = 3.24PSRR973 pKa = 11.84YY974 pKa = 8.21TSPQDD979 pKa = 3.42CPRR982 pKa = 11.84TGDD985 pKa = 3.27RR986 pKa = 11.84QKK988 pKa = 11.33AYY990 pKa = 9.56EE991 pKa = 4.12NTMATYY997 pKa = 10.48ADD999 pKa = 4.49EE1000 pKa = 4.39YY1001 pKa = 10.07AACGSSSSCKK1011 pKa = 10.39NEE1013 pKa = 4.1AKK1015 pKa = 10.45DD1016 pKa = 3.91KK1017 pKa = 10.73RR1018 pKa = 11.84DD1019 pKa = 3.43AKK1021 pKa = 10.71AVEE1024 pKa = 4.25DD1025 pKa = 4.09AKK1027 pKa = 11.52VPLL1030 pKa = 4.25
MM1 pKa = 7.49PRR3 pKa = 11.84QYY5 pKa = 11.43DD6 pKa = 3.87SNDD9 pKa = 3.18VNPFEE14 pKa = 5.81DD15 pKa = 4.38LGPLQFPPTDD25 pKa = 3.68EE26 pKa = 4.71NSPDD30 pKa = 4.23DD31 pKa = 3.89GSLIYY36 pKa = 10.47IHH38 pKa = 7.24GNTTYY43 pKa = 10.25IWDD46 pKa = 3.95GEE48 pKa = 4.21KK49 pKa = 8.34WTASTSGPRR58 pKa = 11.84LVDD61 pKa = 3.59LTWDD65 pKa = 3.93PNSGGGWLLNNAGADD80 pKa = 3.32VWFASVNEE88 pKa = 3.94VDD90 pKa = 4.48AGLMTPDD97 pKa = 2.68ILGRR101 pKa = 11.84IEE103 pKa = 4.48ALEE106 pKa = 4.02LHH108 pKa = 6.75GGAQGIVIKK117 pKa = 10.83GRR119 pKa = 11.84LDD121 pKa = 3.43SVGPPDD127 pKa = 4.1PSLAISVGDD136 pKa = 4.53AYY138 pKa = 11.44LDD140 pKa = 3.75TNGDD144 pKa = 3.24LWVCTEE150 pKa = 3.79EE151 pKa = 5.18LEE153 pKa = 4.35FVNVGMVQGPPGPQGPAGPQGPRR176 pKa = 11.84GYY178 pKa = 10.63DD179 pKa = 3.31GEE181 pKa = 4.68NGLDD185 pKa = 3.76GEE187 pKa = 4.71PGRR190 pKa = 11.84DD191 pKa = 3.65GTPGAQGPPGAPGRR205 pKa = 11.84DD206 pKa = 3.8GQPGAQGEE214 pKa = 4.12QGPQGEE220 pKa = 4.31KK221 pKa = 10.97GEE223 pKa = 4.49RR224 pKa = 11.84GWEE227 pKa = 4.28GATGPQGEE235 pKa = 4.7EE236 pKa = 4.79GPVGPQGPKK245 pKa = 10.04GDD247 pKa = 4.03RR248 pKa = 11.84GDD250 pKa = 3.79VGPEE254 pKa = 4.01GPVVGVEE261 pKa = 4.3GPEE264 pKa = 4.45GPVGPPGPEE273 pKa = 4.11GPEE276 pKa = 4.47GPDD279 pKa = 3.74GPQGPPGTSFANLKK293 pKa = 10.3GVVATEE299 pKa = 4.14HH300 pKa = 7.27DD301 pKa = 4.47LPADD305 pKa = 3.51PTQWDD310 pKa = 3.96MYY312 pKa = 10.59LVEE315 pKa = 4.98SADD318 pKa = 5.39KK319 pKa = 8.94YY320 pKa = 8.95TIWDD324 pKa = 3.77GEE326 pKa = 3.9KK327 pKa = 9.21WRR329 pKa = 11.84YY330 pKa = 7.89VAAGGSVEE338 pKa = 4.82DD339 pKa = 4.74APGDD343 pKa = 3.65SKK345 pKa = 11.5PYY347 pKa = 10.27VRR349 pKa = 11.84YY350 pKa = 10.7NMDD353 pKa = 2.47WRR355 pKa = 11.84EE356 pKa = 4.03GVLDD360 pKa = 4.72APDD363 pKa = 4.33DD364 pKa = 4.0GKK366 pKa = 11.65GYY368 pKa = 10.72VRR370 pKa = 11.84YY371 pKa = 8.28QQGWASLEE379 pKa = 4.21AVGLPYY385 pKa = 10.84LIGTDD390 pKa = 3.22KK391 pKa = 11.03TGLARR396 pKa = 11.84SVRR399 pKa = 11.84KK400 pKa = 9.89AGPFTNTKK408 pKa = 11.0DD409 pKa = 3.32MGPEE413 pKa = 3.45IEE415 pKa = 5.82LIDD418 pKa = 4.46GNGNYY423 pKa = 10.45SNVKK427 pKa = 9.15IGGSGGVVTSSDD439 pKa = 3.24AAGITVDD446 pKa = 4.86GSYY449 pKa = 11.12LLKK452 pKa = 10.7KK453 pKa = 10.57QFDD456 pKa = 4.35EE457 pKa = 4.71YY458 pKa = 10.24PTIDD462 pKa = 4.56AEE464 pKa = 4.53TLDD467 pKa = 4.3SVPADD472 pKa = 2.98SVMFAANQQGNKK484 pKa = 9.93SITWKK489 pKa = 10.8EE490 pKa = 3.7MMGSVMHH497 pKa = 6.24QIGFAIQSASITEE510 pKa = 4.32DD511 pKa = 3.22NAPAQIDD518 pKa = 3.63RR519 pKa = 11.84AVEE522 pKa = 4.36CTCQATLEE530 pKa = 4.27SSAPQFGEE538 pKa = 4.43DD539 pKa = 3.06VCTYY543 pKa = 8.88RR544 pKa = 11.84WRR546 pKa = 11.84AFPQGTSDD554 pKa = 3.32NAKK557 pKa = 10.29PEE559 pKa = 4.09WEE561 pKa = 4.08NGKK564 pKa = 9.66VPSNPNKK571 pKa = 10.05KK572 pKa = 10.41SFVFDD577 pKa = 3.29EE578 pKa = 4.82AGDD581 pKa = 3.77YY582 pKa = 10.57TIEE585 pKa = 4.54CEE587 pKa = 4.3VGASKK592 pKa = 10.89SKK594 pKa = 10.31LWNQVTKK601 pKa = 10.08VASYY605 pKa = 10.91DD606 pKa = 3.44FHH608 pKa = 8.3VKK610 pKa = 10.32EE611 pKa = 4.9GPDD614 pKa = 3.4PLMSYY619 pKa = 10.87AYY621 pKa = 10.32LHH623 pKa = 6.22VKK625 pKa = 10.26NITGGQIKK633 pKa = 9.8FPNALDD639 pKa = 4.1SGNHH643 pKa = 5.37YY644 pKa = 10.72YY645 pKa = 10.69DD646 pKa = 3.76WEE648 pKa = 4.46NMGHH652 pKa = 5.37EE653 pKa = 4.21PEE655 pKa = 4.88YY656 pKa = 10.36YY657 pKa = 10.77AFVHH661 pKa = 5.8VLGLEE666 pKa = 4.53GGDD669 pKa = 3.34QAFGGFKK676 pKa = 10.35CYY678 pKa = 10.56RR679 pKa = 11.84NVQGDD684 pKa = 3.35WDD686 pKa = 3.73DD687 pKa = 3.8WEE689 pKa = 4.46TDD691 pKa = 3.61KK692 pKa = 11.71SDD694 pKa = 3.68EE695 pKa = 4.24TCAKK699 pKa = 10.25YY700 pKa = 10.46GISFYY705 pKa = 11.52DD706 pKa = 3.18NDD708 pKa = 5.1LYY710 pKa = 11.47LEE712 pKa = 4.89PDD714 pKa = 3.0TGLEE718 pKa = 3.86YY719 pKa = 9.88MVYY722 pKa = 9.77MRR724 pKa = 11.84TQTYY728 pKa = 8.81NQVGGSDD735 pKa = 3.66DD736 pKa = 4.01SGVDD740 pKa = 3.54GLNQAGAWLDD750 pKa = 3.52GQRR753 pKa = 11.84WSTTGNATWEE763 pKa = 4.13WGEE766 pKa = 3.92LTDD769 pKa = 3.85TSKK772 pKa = 9.42VTSFYY777 pKa = 11.31RR778 pKa = 11.84MFEE781 pKa = 3.95QFSNSGSAWPTGWEE795 pKa = 4.12HH796 pKa = 7.06FDD798 pKa = 3.2TGKK801 pKa = 11.11GEE803 pKa = 3.8NFAWMFYY810 pKa = 7.6QARR813 pKa = 11.84ATGGIGCGMEE823 pKa = 3.86QWNMSKK829 pKa = 10.31ARR831 pKa = 11.84SLHH834 pKa = 5.48RR835 pKa = 11.84MFYY838 pKa = 9.38QAEE841 pKa = 4.14NFDD844 pKa = 4.53CDD846 pKa = 3.09ISNWDD851 pKa = 3.28TRR853 pKa = 11.84EE854 pKa = 3.84NEE856 pKa = 3.79TLYY859 pKa = 11.19YY860 pKa = 11.0FMGQCKK866 pKa = 10.02NFTCGGLNASEE877 pKa = 4.31EE878 pKa = 5.01GYY880 pKa = 8.66GTGIVNMKK888 pKa = 6.96TLKK891 pKa = 10.54VKK893 pKa = 10.19EE894 pKa = 4.2INYY897 pKa = 9.94SFYY900 pKa = 10.81QCEE903 pKa = 4.26KK904 pKa = 10.93LYY906 pKa = 10.31IDD908 pKa = 4.27CSQWGFPNLEE918 pKa = 4.0NFDD921 pKa = 4.27WDD923 pKa = 3.17TDD925 pKa = 3.73AAPDD929 pKa = 3.5VEE931 pKa = 4.86MFQTANIPRR940 pKa = 11.84SKK942 pKa = 11.01ASDD945 pKa = 3.26YY946 pKa = 10.92WGITSYY952 pKa = 11.52DD953 pKa = 3.84SYY955 pKa = 11.29WNVQIAGILPIWGTDD970 pKa = 3.24PSRR973 pKa = 11.84YY974 pKa = 8.21TSPQDD979 pKa = 3.42CPRR982 pKa = 11.84TGDD985 pKa = 3.27RR986 pKa = 11.84QKK988 pKa = 11.33AYY990 pKa = 9.56EE991 pKa = 4.12NTMATYY997 pKa = 10.48ADD999 pKa = 4.49EE1000 pKa = 4.39YY1001 pKa = 10.07AACGSSSSCKK1011 pKa = 10.39NEE1013 pKa = 4.1AKK1015 pKa = 10.45DD1016 pKa = 3.91KK1017 pKa = 10.73RR1018 pKa = 11.84DD1019 pKa = 3.43AKK1021 pKa = 10.71AVEE1024 pKa = 4.25DD1025 pKa = 4.09AKK1027 pKa = 11.52VPLL1030 pKa = 4.25
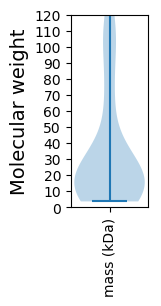
Molecular weight: 112.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6S4PEE1|A0A6S4PEE1_9CAUD Uncharacterized protein OS=uncultured phage_MedDCM-OCT-S38-C3 OX=2740803 PE=4 SV=1
MM1 pKa = 7.65CFGGGGGGGTIVMPNTGAYY20 pKa = 10.96DD21 pKa = 3.39MMANAQISAMQSAMQMGTQRR41 pKa = 11.84VQAQLNDD48 pKa = 3.84SLRR51 pKa = 11.84MQGQYY56 pKa = 7.12QQRR59 pKa = 11.84LRR61 pKa = 11.84DD62 pKa = 3.68IQIEE66 pKa = 4.14RR67 pKa = 11.84AEE69 pKa = 4.12NTQAQARR76 pKa = 11.84RR77 pKa = 11.84MAEE80 pKa = 4.28LIGPPPPEE88 pKa = 4.13EE89 pKa = 3.79TAKK92 pKa = 11.14APGTNEE98 pKa = 3.43KK99 pKa = 9.8TSRR102 pKa = 11.84NNKK105 pKa = 8.8SRR107 pKa = 11.84LRR109 pKa = 11.84INRR112 pKa = 11.84PRR114 pKa = 11.84ATTSAQGTGLNINLGGG130 pKa = 3.61
MM1 pKa = 7.65CFGGGGGGGTIVMPNTGAYY20 pKa = 10.96DD21 pKa = 3.39MMANAQISAMQSAMQMGTQRR41 pKa = 11.84VQAQLNDD48 pKa = 3.84SLRR51 pKa = 11.84MQGQYY56 pKa = 7.12QQRR59 pKa = 11.84LRR61 pKa = 11.84DD62 pKa = 3.68IQIEE66 pKa = 4.14RR67 pKa = 11.84AEE69 pKa = 4.12NTQAQARR76 pKa = 11.84RR77 pKa = 11.84MAEE80 pKa = 4.28LIGPPPPEE88 pKa = 4.13EE89 pKa = 3.79TAKK92 pKa = 11.14APGTNEE98 pKa = 3.43KK99 pKa = 9.8TSRR102 pKa = 11.84NNKK105 pKa = 8.8SRR107 pKa = 11.84LRR109 pKa = 11.84INRR112 pKa = 11.84PRR114 pKa = 11.84ATTSAQGTGLNINLGGG130 pKa = 3.61
Molecular weight: 13.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13510 |
32 |
1124 |
300.2 |
33.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.511 ± 0.396 | 1.088 ± 0.189 |
6.38 ± 0.382 | 6.728 ± 0.236 |
3.124 ± 0.192 | 8.423 ± 0.413 |
1.288 ± 0.209 | 4.367 ± 0.235 |
5.278 ± 0.276 | 8.298 ± 0.432 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.746 ± 0.195 | 4.308 ± 0.213 |
5.056 ± 0.345 | 5.3 ± 0.369 |
5.64 ± 0.466 | 5.759 ± 0.198 |
5.811 ± 0.387 | 6.092 ± 0.224 |
1.991 ± 0.241 | 2.813 ± 0.192 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
