
Muricauda lutaonensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Muricauda
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
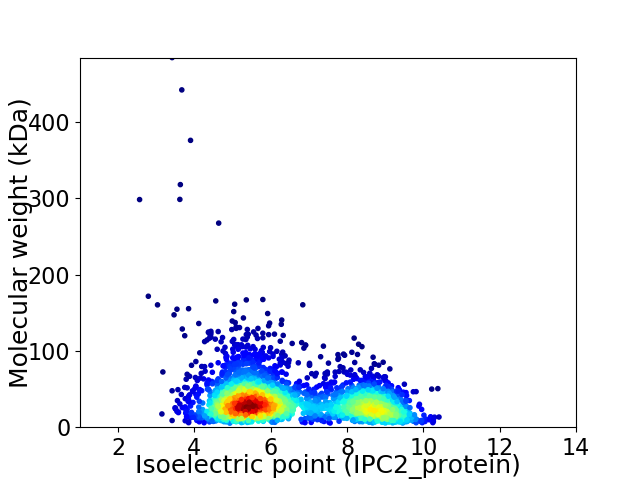
Virtual 2D-PAGE plot for 2882 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D5YVH3|A0A0D5YVH3_9FLAO Uncharacterized protein OS=Muricauda lutaonensis OX=516051 GN=VC82_2257 PE=4 SV=1
MM1 pKa = 7.49GKK3 pKa = 8.33MLKK6 pKa = 9.95IFVLFFLLTISIANAQDD23 pKa = 3.15TYY25 pKa = 11.34YY26 pKa = 10.24DD27 pKa = 3.72TFSSVSYY34 pKa = 10.67SNNDD38 pKa = 2.73GTQNWSTNWIEE49 pKa = 4.62SGDD52 pKa = 4.03DD53 pKa = 3.46NSPSGGYY60 pKa = 9.44IRR62 pKa = 11.84ITGNQLRR69 pKa = 11.84FAYY72 pKa = 9.21IWTEE76 pKa = 3.95TIRR79 pKa = 11.84RR80 pKa = 11.84SADD83 pKa = 2.89LSAYY87 pKa = 10.5SSATLSFDD95 pKa = 2.93WQTSSLEE102 pKa = 4.13SGEE105 pKa = 4.19TLDD108 pKa = 4.32VQVSSDD114 pKa = 3.04GSSYY118 pKa = 7.99TTLATFSGNQSGTFSQNISAHH139 pKa = 5.8ISSNTTIRR147 pKa = 11.84FIKK150 pKa = 10.4GGNNWSGNNDD160 pKa = 2.74RR161 pKa = 11.84VYY163 pKa = 10.71IDD165 pKa = 3.91NVLITATATASVSIADD181 pKa = 3.55VSVNEE186 pKa = 4.27GDD188 pKa = 3.54GTATFMVQHH197 pKa = 6.47VGPNATGAFTVNFTTNDD214 pKa = 3.2GTAVAGSDD222 pKa = 3.56YY223 pKa = 9.81TASSGTLSFNGTSGDD238 pKa = 3.86TEE240 pKa = 5.27SITVPINDD248 pKa = 4.52DD249 pKa = 3.56GDD251 pKa = 4.15FEE253 pKa = 4.46GLEE256 pKa = 4.24TFTVSFTGTSDD267 pKa = 3.23GTINISDD274 pKa = 4.01TATGSITDD282 pKa = 3.94NDD284 pKa = 4.33LLGNTPLALFEE295 pKa = 5.23AFDD298 pKa = 4.64GYY300 pKa = 11.75VDD302 pKa = 3.73YY303 pKa = 10.63TSTGGTLRR311 pKa = 11.84TQPNTGDD318 pKa = 3.02ACAITTSSSNTLTSSIPATATIQKK342 pKa = 10.6ALLYY346 pKa = 7.77WAHH349 pKa = 6.44SAATPDD355 pKa = 3.46SQVTFEE361 pKa = 4.74GNTVTADD368 pKa = 3.47LMYY371 pKa = 7.82TTTLTNRR378 pKa = 11.84VFFGGVSDD386 pKa = 3.7VTSIVQGISNPSTNTYY402 pKa = 11.07DD403 pKa = 3.75FTDD406 pKa = 3.63LTVDD410 pKa = 3.25NTGNYY415 pKa = 9.69CSTATVLGGWSLFIFYY431 pKa = 10.47EE432 pKa = 4.43DD433 pKa = 3.65ASLPASTVNLYY444 pKa = 10.58QGFNGEE450 pKa = 4.72SNSSSSFSLSGFFAIGASGSKK471 pKa = 7.02TTVLSWEE478 pKa = 4.35GDD480 pKa = 3.31QTLSNNEE487 pKa = 4.26LLTVTTGLGTFTLAGDD503 pKa = 3.95GDD505 pKa = 4.49NNGVAVNNPFNSTIFDD521 pKa = 3.34NTIIPNVNNASAYY534 pKa = 10.45GLDD537 pKa = 3.84LDD539 pKa = 4.69TYY541 pKa = 10.88DD542 pKa = 3.65VSPYY546 pKa = 9.34IQPGEE551 pKa = 4.33STVTTNVQSGQDD563 pKa = 3.7FVIMNALVLKK573 pKa = 10.55VPSNLITGTVFEE585 pKa = 4.77DD586 pKa = 3.34VNYY589 pKa = 11.09GGGAGRR595 pKa = 11.84NRR597 pKa = 11.84ASSSGVGVQGATVEE611 pKa = 5.45LYY613 pKa = 10.24DD614 pKa = 4.31ALNNFVDD621 pKa = 4.06NTTTDD626 pKa = 2.97ANGLYY631 pKa = 10.26RR632 pKa = 11.84FGGMANGSYY641 pKa = 9.93SVRR644 pKa = 11.84VVNGTVRR651 pKa = 11.84STRR654 pKa = 11.84GGGAACNTCWPVQTFRR670 pKa = 11.84TSFPGGSVIEE680 pKa = 4.33VTNEE684 pKa = 3.38VGGADD689 pKa = 3.81PSAEE693 pKa = 3.89DD694 pKa = 3.55SAAGTLTGAQSVAPVLITSEE714 pKa = 4.62GVVGMDD720 pKa = 3.81FGYY723 pKa = 11.06NFNTIVNTNEE733 pKa = 4.11DD734 pKa = 3.71GQGSLEE740 pKa = 3.96QFIVNANNLDD750 pKa = 4.17EE751 pKa = 5.45IGLDD755 pKa = 3.1IVANSIFDD763 pKa = 3.84PAAGQDD769 pKa = 3.34TSIFMIPSSSDD780 pKa = 3.21PLGRR784 pKa = 11.84TPDD787 pKa = 3.37SNFASGYY794 pKa = 9.29FDD796 pKa = 4.8IFISDD801 pKa = 4.5GNPLSDD807 pKa = 2.97ITADD811 pKa = 3.19NTFIDD816 pKa = 3.98GRR818 pKa = 11.84TQTAYY823 pKa = 10.84SGDD826 pKa = 4.11TNSGTIGAGGNPVGVSGSLLPIFEE850 pKa = 4.64LPEE853 pKa = 4.12IQVHH857 pKa = 6.33RR858 pKa = 11.84DD859 pKa = 3.0GGDD862 pKa = 3.42VLKK865 pKa = 11.1VQADD869 pKa = 3.65VVLRR873 pKa = 11.84NLSIYY878 pKa = 10.79ADD880 pKa = 3.64NTAGIQVGSGSAVIEE895 pKa = 4.13RR896 pKa = 11.84NLIGVNAQGTNSGNINYY913 pKa = 10.03GIEE916 pKa = 4.3NIGGSILIDD925 pKa = 3.41GNYY928 pKa = 9.66IATNTDD934 pKa = 2.2AGIFVNGGTTNIIQNNHH951 pKa = 4.4ITNNGNSACTDD962 pKa = 3.63NILVQGGSGLVITQNFIEE980 pKa = 4.47NAASAGIDD988 pKa = 3.28ASASSGNITISEE1000 pKa = 4.06NSITASGQDD1009 pKa = 3.42GGNCSGNIEE1018 pKa = 3.98GMAIKK1023 pKa = 10.26IGGNNSVVTNNMIYY1037 pKa = 10.77ANGGAGIVVVGGTANRR1053 pKa = 11.84ISQNSIYY1060 pKa = 11.06ANGTISDD1067 pKa = 4.14ALGIDD1072 pKa = 4.48LDD1074 pKa = 4.2ASSGAGDD1081 pKa = 4.12GVTLNDD1087 pKa = 4.52NGDD1090 pKa = 3.74GDD1092 pKa = 4.15TGPNDD1097 pKa = 3.7LQNFPIISAAYY1108 pKa = 8.58ISGSNLVVKK1117 pKa = 10.34GWSRR1121 pKa = 11.84PGATIEE1127 pKa = 5.38FFFTDD1132 pKa = 3.6INEE1135 pKa = 4.34GTATTGDD1142 pKa = 3.83NQLGMAVDD1150 pKa = 3.8YY1151 pKa = 11.67GEE1153 pKa = 4.39GQTFIGAAVEE1163 pKa = 4.33GSGADD1168 pKa = 3.46ADD1170 pKa = 4.29NTSSSYY1176 pKa = 11.66SDD1178 pKa = 3.31VDD1180 pKa = 3.85GNTDD1184 pKa = 2.99NTNRR1188 pKa = 11.84FHH1190 pKa = 7.56FNLPVPSGATLGEE1203 pKa = 4.73MITATATLGNSTSEE1217 pKa = 4.18FAPMSILRR1225 pKa = 11.84AATVITNRR1233 pKa = 11.84RR1234 pKa = 11.84ITYY1237 pKa = 9.66RR1238 pKa = 11.84VSKK1241 pKa = 10.76NN1242 pKa = 3.0
MM1 pKa = 7.49GKK3 pKa = 8.33MLKK6 pKa = 9.95IFVLFFLLTISIANAQDD23 pKa = 3.15TYY25 pKa = 11.34YY26 pKa = 10.24DD27 pKa = 3.72TFSSVSYY34 pKa = 10.67SNNDD38 pKa = 2.73GTQNWSTNWIEE49 pKa = 4.62SGDD52 pKa = 4.03DD53 pKa = 3.46NSPSGGYY60 pKa = 9.44IRR62 pKa = 11.84ITGNQLRR69 pKa = 11.84FAYY72 pKa = 9.21IWTEE76 pKa = 3.95TIRR79 pKa = 11.84RR80 pKa = 11.84SADD83 pKa = 2.89LSAYY87 pKa = 10.5SSATLSFDD95 pKa = 2.93WQTSSLEE102 pKa = 4.13SGEE105 pKa = 4.19TLDD108 pKa = 4.32VQVSSDD114 pKa = 3.04GSSYY118 pKa = 7.99TTLATFSGNQSGTFSQNISAHH139 pKa = 5.8ISSNTTIRR147 pKa = 11.84FIKK150 pKa = 10.4GGNNWSGNNDD160 pKa = 2.74RR161 pKa = 11.84VYY163 pKa = 10.71IDD165 pKa = 3.91NVLITATATASVSIADD181 pKa = 3.55VSVNEE186 pKa = 4.27GDD188 pKa = 3.54GTATFMVQHH197 pKa = 6.47VGPNATGAFTVNFTTNDD214 pKa = 3.2GTAVAGSDD222 pKa = 3.56YY223 pKa = 9.81TASSGTLSFNGTSGDD238 pKa = 3.86TEE240 pKa = 5.27SITVPINDD248 pKa = 4.52DD249 pKa = 3.56GDD251 pKa = 4.15FEE253 pKa = 4.46GLEE256 pKa = 4.24TFTVSFTGTSDD267 pKa = 3.23GTINISDD274 pKa = 4.01TATGSITDD282 pKa = 3.94NDD284 pKa = 4.33LLGNTPLALFEE295 pKa = 5.23AFDD298 pKa = 4.64GYY300 pKa = 11.75VDD302 pKa = 3.73YY303 pKa = 10.63TSTGGTLRR311 pKa = 11.84TQPNTGDD318 pKa = 3.02ACAITTSSSNTLTSSIPATATIQKK342 pKa = 10.6ALLYY346 pKa = 7.77WAHH349 pKa = 6.44SAATPDD355 pKa = 3.46SQVTFEE361 pKa = 4.74GNTVTADD368 pKa = 3.47LMYY371 pKa = 7.82TTTLTNRR378 pKa = 11.84VFFGGVSDD386 pKa = 3.7VTSIVQGISNPSTNTYY402 pKa = 11.07DD403 pKa = 3.75FTDD406 pKa = 3.63LTVDD410 pKa = 3.25NTGNYY415 pKa = 9.69CSTATVLGGWSLFIFYY431 pKa = 10.47EE432 pKa = 4.43DD433 pKa = 3.65ASLPASTVNLYY444 pKa = 10.58QGFNGEE450 pKa = 4.72SNSSSSFSLSGFFAIGASGSKK471 pKa = 7.02TTVLSWEE478 pKa = 4.35GDD480 pKa = 3.31QTLSNNEE487 pKa = 4.26LLTVTTGLGTFTLAGDD503 pKa = 3.95GDD505 pKa = 4.49NNGVAVNNPFNSTIFDD521 pKa = 3.34NTIIPNVNNASAYY534 pKa = 10.45GLDD537 pKa = 3.84LDD539 pKa = 4.69TYY541 pKa = 10.88DD542 pKa = 3.65VSPYY546 pKa = 9.34IQPGEE551 pKa = 4.33STVTTNVQSGQDD563 pKa = 3.7FVIMNALVLKK573 pKa = 10.55VPSNLITGTVFEE585 pKa = 4.77DD586 pKa = 3.34VNYY589 pKa = 11.09GGGAGRR595 pKa = 11.84NRR597 pKa = 11.84ASSSGVGVQGATVEE611 pKa = 5.45LYY613 pKa = 10.24DD614 pKa = 4.31ALNNFVDD621 pKa = 4.06NTTTDD626 pKa = 2.97ANGLYY631 pKa = 10.26RR632 pKa = 11.84FGGMANGSYY641 pKa = 9.93SVRR644 pKa = 11.84VVNGTVRR651 pKa = 11.84STRR654 pKa = 11.84GGGAACNTCWPVQTFRR670 pKa = 11.84TSFPGGSVIEE680 pKa = 4.33VTNEE684 pKa = 3.38VGGADD689 pKa = 3.81PSAEE693 pKa = 3.89DD694 pKa = 3.55SAAGTLTGAQSVAPVLITSEE714 pKa = 4.62GVVGMDD720 pKa = 3.81FGYY723 pKa = 11.06NFNTIVNTNEE733 pKa = 4.11DD734 pKa = 3.71GQGSLEE740 pKa = 3.96QFIVNANNLDD750 pKa = 4.17EE751 pKa = 5.45IGLDD755 pKa = 3.1IVANSIFDD763 pKa = 3.84PAAGQDD769 pKa = 3.34TSIFMIPSSSDD780 pKa = 3.21PLGRR784 pKa = 11.84TPDD787 pKa = 3.37SNFASGYY794 pKa = 9.29FDD796 pKa = 4.8IFISDD801 pKa = 4.5GNPLSDD807 pKa = 2.97ITADD811 pKa = 3.19NTFIDD816 pKa = 3.98GRR818 pKa = 11.84TQTAYY823 pKa = 10.84SGDD826 pKa = 4.11TNSGTIGAGGNPVGVSGSLLPIFEE850 pKa = 4.64LPEE853 pKa = 4.12IQVHH857 pKa = 6.33RR858 pKa = 11.84DD859 pKa = 3.0GGDD862 pKa = 3.42VLKK865 pKa = 11.1VQADD869 pKa = 3.65VVLRR873 pKa = 11.84NLSIYY878 pKa = 10.79ADD880 pKa = 3.64NTAGIQVGSGSAVIEE895 pKa = 4.13RR896 pKa = 11.84NLIGVNAQGTNSGNINYY913 pKa = 10.03GIEE916 pKa = 4.3NIGGSILIDD925 pKa = 3.41GNYY928 pKa = 9.66IATNTDD934 pKa = 2.2AGIFVNGGTTNIIQNNHH951 pKa = 4.4ITNNGNSACTDD962 pKa = 3.63NILVQGGSGLVITQNFIEE980 pKa = 4.47NAASAGIDD988 pKa = 3.28ASASSGNITISEE1000 pKa = 4.06NSITASGQDD1009 pKa = 3.42GGNCSGNIEE1018 pKa = 3.98GMAIKK1023 pKa = 10.26IGGNNSVVTNNMIYY1037 pKa = 10.77ANGGAGIVVVGGTANRR1053 pKa = 11.84ISQNSIYY1060 pKa = 11.06ANGTISDD1067 pKa = 4.14ALGIDD1072 pKa = 4.48LDD1074 pKa = 4.2ASSGAGDD1081 pKa = 4.12GVTLNDD1087 pKa = 4.52NGDD1090 pKa = 3.74GDD1092 pKa = 4.15TGPNDD1097 pKa = 3.7LQNFPIISAAYY1108 pKa = 8.58ISGSNLVVKK1117 pKa = 10.34GWSRR1121 pKa = 11.84PGATIEE1127 pKa = 5.38FFFTDD1132 pKa = 3.6INEE1135 pKa = 4.34GTATTGDD1142 pKa = 3.83NQLGMAVDD1150 pKa = 3.8YY1151 pKa = 11.67GEE1153 pKa = 4.39GQTFIGAAVEE1163 pKa = 4.33GSGADD1168 pKa = 3.46ADD1170 pKa = 4.29NTSSSYY1176 pKa = 11.66SDD1178 pKa = 3.31VDD1180 pKa = 3.85GNTDD1184 pKa = 2.99NTNRR1188 pKa = 11.84FHH1190 pKa = 7.56FNLPVPSGATLGEE1203 pKa = 4.73MITATATLGNSTSEE1217 pKa = 4.18FAPMSILRR1225 pKa = 11.84AATVITNRR1233 pKa = 11.84RR1234 pKa = 11.84ITYY1237 pKa = 9.66RR1238 pKa = 11.84VSKK1241 pKa = 10.76NN1242 pKa = 3.0
Molecular weight: 128.63 kDa
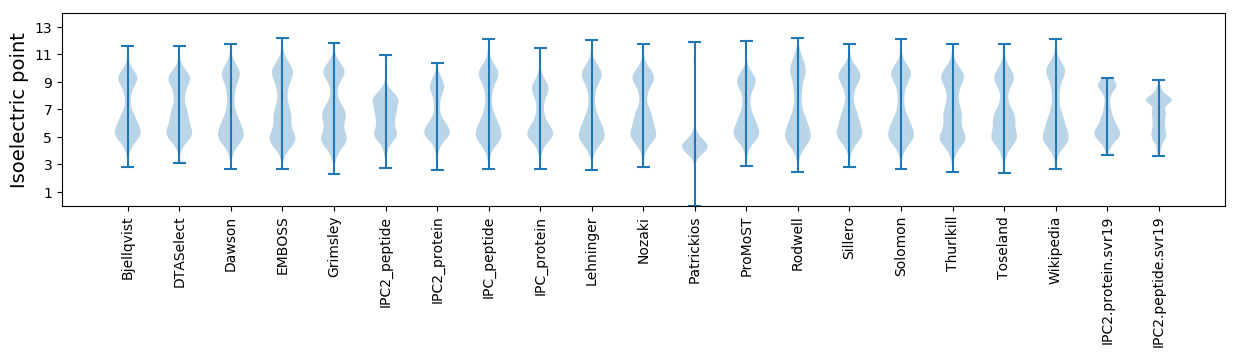
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D5YRP8|A0A0D5YRP8_9FLAO Uncharacterized protein OS=Muricauda lutaonensis OX=516051 GN=VC82_1328 PE=4 SV=1
MM1 pKa = 7.46NEE3 pKa = 3.5VSLLDD8 pKa = 3.32WFLAFTAIFLLGISKK23 pKa = 10.51SGLKK27 pKa = 10.23GLSIFNVTLLALAFGARR44 pKa = 11.84EE45 pKa = 4.05STGILIPLLLVGDD58 pKa = 4.35VFAVLYY64 pKa = 9.15YY65 pKa = 10.44NRR67 pKa = 11.84HH68 pKa = 4.39TQWRR72 pKa = 11.84YY73 pKa = 9.19ILRR76 pKa = 11.84FLPWMISGILIGVFLGKK93 pKa = 9.89NLPEE97 pKa = 4.12TQFKK101 pKa = 10.11IGMVIVIFISLGVLTYY117 pKa = 9.61WDD119 pKa = 3.4RR120 pKa = 11.84RR121 pKa = 11.84KK122 pKa = 10.3SKK124 pKa = 10.37KK125 pKa = 10.44VPTHH129 pKa = 4.16WTFAGGVGILAGICTMIGNLAGAFTNIFFLAMRR162 pKa = 11.84LPKK165 pKa = 10.57NEE167 pKa = 4.14FVGTAAWLFLITNLFKK183 pKa = 11.08LPFHH187 pKa = 5.87IWVWKK192 pKa = 8.77TITPEE197 pKa = 3.93TVGINLKK204 pKa = 10.0LLPAILVGLAIGVRR218 pKa = 11.84IVRR221 pKa = 11.84RR222 pKa = 11.84INEE225 pKa = 3.62KK226 pKa = 8.96QYY228 pKa = 10.98RR229 pKa = 11.84RR230 pKa = 11.84FILIVTAIGAVVILLRR246 pKa = 4.58
MM1 pKa = 7.46NEE3 pKa = 3.5VSLLDD8 pKa = 3.32WFLAFTAIFLLGISKK23 pKa = 10.51SGLKK27 pKa = 10.23GLSIFNVTLLALAFGARR44 pKa = 11.84EE45 pKa = 4.05STGILIPLLLVGDD58 pKa = 4.35VFAVLYY64 pKa = 9.15YY65 pKa = 10.44NRR67 pKa = 11.84HH68 pKa = 4.39TQWRR72 pKa = 11.84YY73 pKa = 9.19ILRR76 pKa = 11.84FLPWMISGILIGVFLGKK93 pKa = 9.89NLPEE97 pKa = 4.12TQFKK101 pKa = 10.11IGMVIVIFISLGVLTYY117 pKa = 9.61WDD119 pKa = 3.4RR120 pKa = 11.84RR121 pKa = 11.84KK122 pKa = 10.3SKK124 pKa = 10.37KK125 pKa = 10.44VPTHH129 pKa = 4.16WTFAGGVGILAGICTMIGNLAGAFTNIFFLAMRR162 pKa = 11.84LPKK165 pKa = 10.57NEE167 pKa = 4.14FVGTAAWLFLITNLFKK183 pKa = 11.08LPFHH187 pKa = 5.87IWVWKK192 pKa = 8.77TITPEE197 pKa = 3.93TVGINLKK204 pKa = 10.0LLPAILVGLAIGVRR218 pKa = 11.84IVRR221 pKa = 11.84RR222 pKa = 11.84INEE225 pKa = 3.62KK226 pKa = 8.96QYY228 pKa = 10.98RR229 pKa = 11.84RR230 pKa = 11.84FILIVTAIGAVVILLRR246 pKa = 4.58
Molecular weight: 27.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
971754 |
50 |
4585 |
337.2 |
37.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.973 ± 0.047 | 0.746 ± 0.017 |
5.839 ± 0.055 | 6.859 ± 0.043 |
5.143 ± 0.037 | 6.989 ± 0.053 |
1.954 ± 0.03 | 6.91 ± 0.041 |
7.198 ± 0.079 | 9.491 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.324 ± 0.028 | 5.332 ± 0.049 |
3.748 ± 0.028 | 3.475 ± 0.026 |
4.09 ± 0.038 | 5.798 ± 0.036 |
5.508 ± 0.066 | 6.663 ± 0.034 |
1.125 ± 0.019 | 3.832 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
