
Corynebacterium urealyticum (strain ATCC 43042 / DSM 7109)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Corynebacteriaceae; Corynebacterium; Corynebacterium urealyticum
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
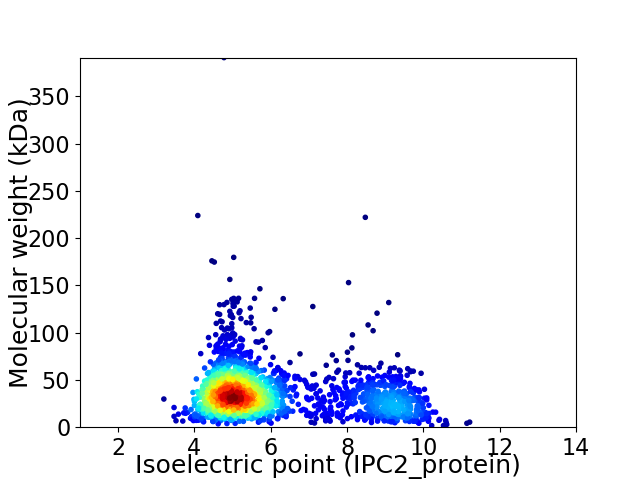
Virtual 2D-PAGE plot for 2011 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B1VGL6|B1VGL6_CORU7 Probable nicotinate-nucleotide adenylyltransferase OS=Corynebacterium urealyticum (strain ATCC 43042 / DSM 7109) OX=504474 GN=nadD PE=3 SV=1
MM1 pKa = 7.02VLSAACANFRR11 pKa = 11.84GMTKK15 pKa = 10.06PQDD18 pKa = 3.37HH19 pKa = 7.0HH20 pKa = 7.19PDD22 pKa = 3.31DD23 pKa = 4.3TGAEE27 pKa = 4.3GGDD30 pKa = 3.37ASGPQPPQYY39 pKa = 9.84RR40 pKa = 11.84RR41 pKa = 11.84PGDD44 pKa = 3.47TPTADD49 pKa = 3.24GSSQQGANPWAQPGAGPSAQQGAGQWGSQGSGQWGQAGAGYY90 pKa = 9.95GAGASQQWAQQNPYY104 pKa = 9.15GAQGQSAQWQGQDD117 pKa = 3.87YY118 pKa = 9.71GAQGYY123 pKa = 9.37NNQGYY128 pKa = 10.14GGPNYY133 pKa = 10.34GNQGYY138 pKa = 8.98PEE140 pKa = 3.98QGYY143 pKa = 10.47AGPGGQWQGQGYY155 pKa = 9.15EE156 pKa = 3.95NQGYY160 pKa = 8.42QEE162 pKa = 4.05QPEE165 pKa = 4.36EE166 pKa = 4.22KK167 pKa = 10.49SNGRR171 pKa = 11.84NILIIVLLVLALLAAVAFAVFWFTGKK197 pKa = 9.7PGEE200 pKa = 4.61GKK202 pKa = 10.34SSEE205 pKa = 4.1QTTVSSSEE213 pKa = 3.84ATDD216 pKa = 3.3ARR218 pKa = 11.84GTEE221 pKa = 4.63DD222 pKa = 4.22EE223 pKa = 5.12DD224 pKa = 5.98ADD226 pKa = 3.94AQGDD230 pKa = 4.7DD231 pKa = 4.03RR232 pKa = 11.84DD233 pKa = 3.63EE234 pKa = 4.45DD235 pKa = 4.12ANRR238 pKa = 11.84EE239 pKa = 4.02DD240 pKa = 5.32DD241 pKa = 5.94DD242 pKa = 5.3EE243 pKa = 4.58DD244 pKa = 3.84TGDD247 pKa = 3.75EE248 pKa = 4.35AEE250 pKa = 4.75SRR252 pKa = 11.84PKK254 pKa = 10.63NPRR257 pKa = 11.84LPGYY261 pKa = 7.35ATPVNSSARR270 pKa = 11.84GSNPGGDD277 pKa = 4.21LNSVWKK283 pKa = 10.55SGPTSDD289 pKa = 4.78SFALAVRR296 pKa = 11.84DD297 pKa = 4.26AYY299 pKa = 11.33VDD301 pKa = 3.68AYY303 pKa = 11.42LDD305 pKa = 3.81GDD307 pKa = 3.58EE308 pKa = 6.08DD309 pKa = 4.01EE310 pKa = 5.07FNHH313 pKa = 5.83VVEE316 pKa = 5.43AYY318 pKa = 10.8SDD320 pKa = 3.79VTGTSYY326 pKa = 11.02TMSCRR331 pKa = 11.84DD332 pKa = 3.09NGEE335 pKa = 4.29YY336 pKa = 9.8VHH338 pKa = 6.46CTGGNDD344 pKa = 3.39ANVYY348 pKa = 9.43IAA350 pKa = 5.1
MM1 pKa = 7.02VLSAACANFRR11 pKa = 11.84GMTKK15 pKa = 10.06PQDD18 pKa = 3.37HH19 pKa = 7.0HH20 pKa = 7.19PDD22 pKa = 3.31DD23 pKa = 4.3TGAEE27 pKa = 4.3GGDD30 pKa = 3.37ASGPQPPQYY39 pKa = 9.84RR40 pKa = 11.84RR41 pKa = 11.84PGDD44 pKa = 3.47TPTADD49 pKa = 3.24GSSQQGANPWAQPGAGPSAQQGAGQWGSQGSGQWGQAGAGYY90 pKa = 9.95GAGASQQWAQQNPYY104 pKa = 9.15GAQGQSAQWQGQDD117 pKa = 3.87YY118 pKa = 9.71GAQGYY123 pKa = 9.37NNQGYY128 pKa = 10.14GGPNYY133 pKa = 10.34GNQGYY138 pKa = 8.98PEE140 pKa = 3.98QGYY143 pKa = 10.47AGPGGQWQGQGYY155 pKa = 9.15EE156 pKa = 3.95NQGYY160 pKa = 8.42QEE162 pKa = 4.05QPEE165 pKa = 4.36EE166 pKa = 4.22KK167 pKa = 10.49SNGRR171 pKa = 11.84NILIIVLLVLALLAAVAFAVFWFTGKK197 pKa = 9.7PGEE200 pKa = 4.61GKK202 pKa = 10.34SSEE205 pKa = 4.1QTTVSSSEE213 pKa = 3.84ATDD216 pKa = 3.3ARR218 pKa = 11.84GTEE221 pKa = 4.63DD222 pKa = 4.22EE223 pKa = 5.12DD224 pKa = 5.98ADD226 pKa = 3.94AQGDD230 pKa = 4.7DD231 pKa = 4.03RR232 pKa = 11.84DD233 pKa = 3.63EE234 pKa = 4.45DD235 pKa = 4.12ANRR238 pKa = 11.84EE239 pKa = 4.02DD240 pKa = 5.32DD241 pKa = 5.94DD242 pKa = 5.3EE243 pKa = 4.58DD244 pKa = 3.84TGDD247 pKa = 3.75EE248 pKa = 4.35AEE250 pKa = 4.75SRR252 pKa = 11.84PKK254 pKa = 10.63NPRR257 pKa = 11.84LPGYY261 pKa = 7.35ATPVNSSARR270 pKa = 11.84GSNPGGDD277 pKa = 4.21LNSVWKK283 pKa = 10.55SGPTSDD289 pKa = 4.78SFALAVRR296 pKa = 11.84DD297 pKa = 4.26AYY299 pKa = 11.33VDD301 pKa = 3.68AYY303 pKa = 11.42LDD305 pKa = 3.81GDD307 pKa = 3.58EE308 pKa = 6.08DD309 pKa = 4.01EE310 pKa = 5.07FNHH313 pKa = 5.83VVEE316 pKa = 5.43AYY318 pKa = 10.8SDD320 pKa = 3.79VTGTSYY326 pKa = 11.02TMSCRR331 pKa = 11.84DD332 pKa = 3.09NGEE335 pKa = 4.29YY336 pKa = 9.8VHH338 pKa = 6.46CTGGNDD344 pKa = 3.39ANVYY348 pKa = 9.43IAA350 pKa = 5.1
Molecular weight: 36.78 kDa
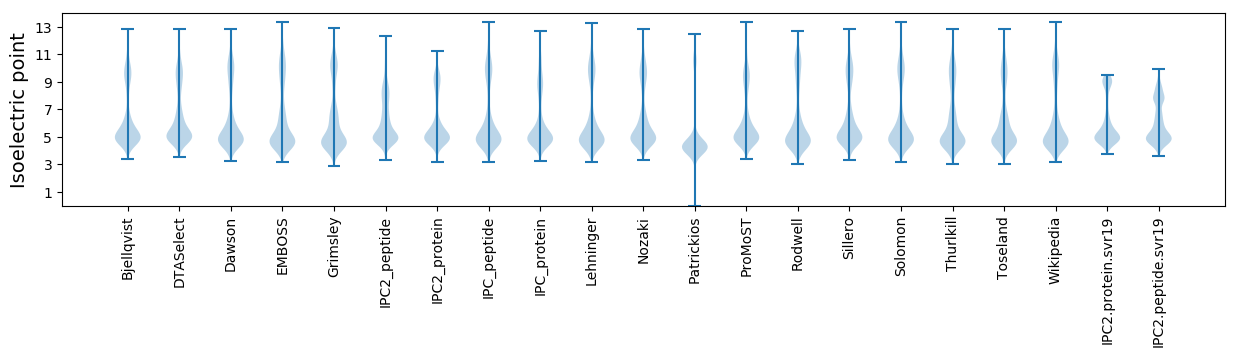
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B1VEK0|B1VEK0_CORU7 Delta-aminolevulinic acid dehydratase OS=Corynebacterium urealyticum (strain ATCC 43042 / DSM 7109) OX=504474 GN=cu0229 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.63KK15 pKa = 9.56KK16 pKa = 9.27HH17 pKa = 5.36RR18 pKa = 11.84KK19 pKa = 5.5MLKK22 pKa = 7.41RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.63KK15 pKa = 9.56KK16 pKa = 9.27HH17 pKa = 5.36RR18 pKa = 11.84KK19 pKa = 5.5MLKK22 pKa = 7.41RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
Molecular weight: 4.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
702396 |
15 |
3618 |
349.3 |
37.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.974 ± 0.077 | 0.662 ± 0.015 |
5.854 ± 0.046 | 6.654 ± 0.056 |
3.123 ± 0.03 | 8.76 ± 0.05 |
2.1 ± 0.023 | 4.687 ± 0.037 |
3.461 ± 0.048 | 9.386 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.184 ± 0.025 | 2.871 ± 0.033 |
5.245 ± 0.045 | 3.618 ± 0.03 |
6.458 ± 0.05 | 5.743 ± 0.039 |
5.848 ± 0.038 | 7.873 ± 0.048 |
1.404 ± 0.022 | 2.094 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
