
Saliniradius amylolyticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Saliniradius
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
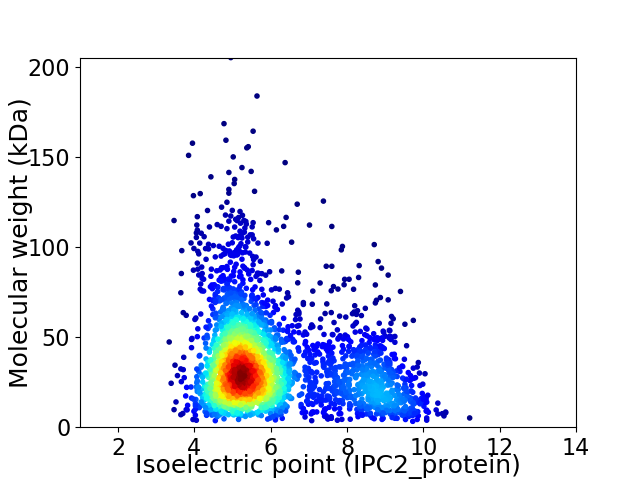
Virtual 2D-PAGE plot for 2945 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
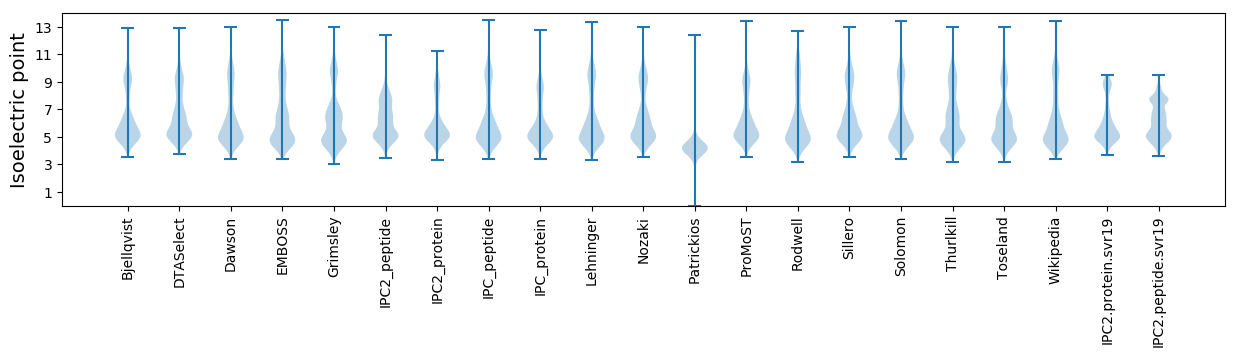
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S2E1X4|A0A2S2E1X4_9ALTE tRNA pseudouridine synthase D OS=Saliniradius amylolyticus OX=2183582 GN=truD PE=3 SV=1
MM1 pKa = 7.79KK2 pKa = 10.42SITPILAALTLPFVLTGCLNSDD24 pKa = 3.81SDD26 pKa = 4.59PLPEE30 pKa = 4.82PPEE33 pKa = 4.15PPAFSNLRR41 pKa = 11.84VTHH44 pKa = 6.33AVSDD48 pKa = 3.96APTVNVIANGDD59 pKa = 4.05TLSEE63 pKa = 4.22GADD66 pKa = 3.74YY67 pKa = 10.63QASTGWQSVLSDD79 pKa = 3.86TYY81 pKa = 10.32TVEE84 pKa = 4.27VNANLPGGDD93 pKa = 3.55SATVIGPVDD102 pKa = 3.77LTLTGDD108 pKa = 3.31MNYY111 pKa = 10.86DD112 pKa = 3.41VVAVGQVGDD121 pKa = 3.93IEE123 pKa = 4.38PLVLEE128 pKa = 4.29NAEE131 pKa = 4.31TTVTSGNARR140 pKa = 11.84VQILHH145 pKa = 6.81AAANAPMVDD154 pKa = 3.63IYY156 pKa = 10.59VTAPDD161 pKa = 4.11AEE163 pKa = 4.48LASEE167 pKa = 4.07QALATLSFKK176 pKa = 10.72EE177 pKa = 4.1YY178 pKa = 9.37TAQTEE183 pKa = 4.58VAGGDD188 pKa = 3.32YY189 pKa = 10.17RR190 pKa = 11.84VRR192 pKa = 11.84ITPAGADD199 pKa = 3.1TVVFDD204 pKa = 4.87SGTINLADD212 pKa = 4.27GADD215 pKa = 3.73LMVAATDD222 pKa = 3.5NTGAGDD228 pKa = 3.99SPVTLLAVDD237 pKa = 3.97ANGATRR243 pKa = 11.84LWDD246 pKa = 3.7TNTGANLRR254 pKa = 11.84VLHH257 pKa = 6.52GSPDD261 pKa = 3.44APAVDD266 pKa = 3.46VLVNNATMPMHH277 pKa = 5.85YY278 pKa = 9.99QLDD281 pKa = 3.73GLAYY285 pKa = 9.81PNATDD290 pKa = 3.9YY291 pKa = 11.51LAVPADD297 pKa = 3.99EE298 pKa = 5.12LLVDD302 pKa = 4.25VVADD306 pKa = 3.74ADD308 pKa = 3.91NSVVAIDD315 pKa = 4.33DD316 pKa = 3.87AAVAPMQGMAYY327 pKa = 8.59TAIATNTLADD337 pKa = 3.52IQLALLTDD345 pKa = 3.72DD346 pKa = 4.43HH347 pKa = 7.79RR348 pKa = 11.84SIATAAKK355 pKa = 9.92VRR357 pKa = 11.84LIHH360 pKa = 6.13NAPAAGDD367 pKa = 3.29VDD369 pKa = 3.95IYY371 pKa = 10.52VTADD375 pKa = 3.79DD376 pKa = 5.98DD377 pKa = 5.31ISDD380 pKa = 5.09DD381 pKa = 4.48DD382 pKa = 4.34PAFTDD387 pKa = 3.6VPYY390 pKa = 10.91DD391 pKa = 3.59ATNLATTGYY400 pKa = 10.37VEE402 pKa = 4.27LAEE405 pKa = 4.03GTYY408 pKa = 10.73YY409 pKa = 9.31ITITPADD416 pKa = 4.06TKK418 pKa = 10.76TEE420 pKa = 4.68AIGPLMVEE428 pKa = 4.59LNNGDD433 pKa = 3.76VLSAVALEE441 pKa = 4.32ANGGGTPLQAVILDD455 pKa = 3.93DD456 pKa = 4.33SKK458 pKa = 10.97PAPVSLDD465 pKa = 3.02ATQTFEE471 pKa = 4.04VSLDD475 pKa = 3.47GMQEE479 pKa = 3.94VPAVDD484 pKa = 3.85TAATAAATVLLDD496 pKa = 3.6EE497 pKa = 5.44DD498 pKa = 4.66DD499 pKa = 4.66RR500 pKa = 11.84LFSVTVDD507 pKa = 3.21TSAVDD512 pKa = 3.39NVTGVHH518 pKa = 4.92VHH520 pKa = 7.22DD521 pKa = 5.15GNIGRR526 pKa = 11.84NGPVAFPLQASGDD539 pKa = 3.8DD540 pKa = 4.3TYY542 pKa = 11.62TLPATNLLDD551 pKa = 4.22PMIEE555 pKa = 3.97ALKK558 pKa = 10.59SGEE561 pKa = 3.88WYY563 pKa = 10.22INVHH567 pKa = 5.27TEE569 pKa = 3.88ANPSGEE575 pKa = 4.01VRR577 pKa = 11.84GQIVPEE583 pKa = 4.05TMAVVTFPLSGSQEE597 pKa = 4.35VPSVTTDD604 pKa = 2.63AMGYY608 pKa = 10.59GYY610 pKa = 9.02ATLDD614 pKa = 3.55TTTLAVDD621 pKa = 4.09LVAVTMGVEE630 pKa = 4.34DD631 pKa = 3.3ATMAHH636 pKa = 6.57IHH638 pKa = 5.92TGYY641 pKa = 10.6AGEE644 pKa = 4.34NGPVYY649 pKa = 9.8VTLEE653 pKa = 3.81QSMDD657 pKa = 3.82DD658 pKa = 3.59ANVWMTPAGAMIDD671 pKa = 3.54ATNAARR677 pKa = 11.84LVEE680 pKa = 4.37GGHH683 pKa = 5.74YY684 pKa = 10.93VNIHH688 pKa = 5.45TPANPSGEE696 pKa = 4.24LRR698 pKa = 11.84GQITPDD704 pKa = 3.38NIEE707 pKa = 4.21VYY709 pKa = 10.57GVTATGDD716 pKa = 3.59QEE718 pKa = 4.69VPAVDD723 pKa = 3.78TNAMGEE729 pKa = 4.25GAITLNTSTMTIKK742 pKa = 10.87AIINVMDD749 pKa = 4.07ISPNAAHH756 pKa = 7.0IHH758 pKa = 5.03QGAVGEE764 pKa = 4.21NGGVVIGLTNPDD776 pKa = 3.33AGLWTLDD783 pKa = 3.25ATLTEE788 pKa = 4.16AQMALMQAEE797 pKa = 4.01GLYY800 pKa = 10.82TNFHH804 pKa = 6.36TDD806 pKa = 3.12AFPSGEE812 pKa = 3.71IRR814 pKa = 11.84GQITLGFEE822 pKa = 4.07
MM1 pKa = 7.79KK2 pKa = 10.42SITPILAALTLPFVLTGCLNSDD24 pKa = 3.81SDD26 pKa = 4.59PLPEE30 pKa = 4.82PPEE33 pKa = 4.15PPAFSNLRR41 pKa = 11.84VTHH44 pKa = 6.33AVSDD48 pKa = 3.96APTVNVIANGDD59 pKa = 4.05TLSEE63 pKa = 4.22GADD66 pKa = 3.74YY67 pKa = 10.63QASTGWQSVLSDD79 pKa = 3.86TYY81 pKa = 10.32TVEE84 pKa = 4.27VNANLPGGDD93 pKa = 3.55SATVIGPVDD102 pKa = 3.77LTLTGDD108 pKa = 3.31MNYY111 pKa = 10.86DD112 pKa = 3.41VVAVGQVGDD121 pKa = 3.93IEE123 pKa = 4.38PLVLEE128 pKa = 4.29NAEE131 pKa = 4.31TTVTSGNARR140 pKa = 11.84VQILHH145 pKa = 6.81AAANAPMVDD154 pKa = 3.63IYY156 pKa = 10.59VTAPDD161 pKa = 4.11AEE163 pKa = 4.48LASEE167 pKa = 4.07QALATLSFKK176 pKa = 10.72EE177 pKa = 4.1YY178 pKa = 9.37TAQTEE183 pKa = 4.58VAGGDD188 pKa = 3.32YY189 pKa = 10.17RR190 pKa = 11.84VRR192 pKa = 11.84ITPAGADD199 pKa = 3.1TVVFDD204 pKa = 4.87SGTINLADD212 pKa = 4.27GADD215 pKa = 3.73LMVAATDD222 pKa = 3.5NTGAGDD228 pKa = 3.99SPVTLLAVDD237 pKa = 3.97ANGATRR243 pKa = 11.84LWDD246 pKa = 3.7TNTGANLRR254 pKa = 11.84VLHH257 pKa = 6.52GSPDD261 pKa = 3.44APAVDD266 pKa = 3.46VLVNNATMPMHH277 pKa = 5.85YY278 pKa = 9.99QLDD281 pKa = 3.73GLAYY285 pKa = 9.81PNATDD290 pKa = 3.9YY291 pKa = 11.51LAVPADD297 pKa = 3.99EE298 pKa = 5.12LLVDD302 pKa = 4.25VVADD306 pKa = 3.74ADD308 pKa = 3.91NSVVAIDD315 pKa = 4.33DD316 pKa = 3.87AAVAPMQGMAYY327 pKa = 8.59TAIATNTLADD337 pKa = 3.52IQLALLTDD345 pKa = 3.72DD346 pKa = 4.43HH347 pKa = 7.79RR348 pKa = 11.84SIATAAKK355 pKa = 9.92VRR357 pKa = 11.84LIHH360 pKa = 6.13NAPAAGDD367 pKa = 3.29VDD369 pKa = 3.95IYY371 pKa = 10.52VTADD375 pKa = 3.79DD376 pKa = 5.98DD377 pKa = 5.31ISDD380 pKa = 5.09DD381 pKa = 4.48DD382 pKa = 4.34PAFTDD387 pKa = 3.6VPYY390 pKa = 10.91DD391 pKa = 3.59ATNLATTGYY400 pKa = 10.37VEE402 pKa = 4.27LAEE405 pKa = 4.03GTYY408 pKa = 10.73YY409 pKa = 9.31ITITPADD416 pKa = 4.06TKK418 pKa = 10.76TEE420 pKa = 4.68AIGPLMVEE428 pKa = 4.59LNNGDD433 pKa = 3.76VLSAVALEE441 pKa = 4.32ANGGGTPLQAVILDD455 pKa = 3.93DD456 pKa = 4.33SKK458 pKa = 10.97PAPVSLDD465 pKa = 3.02ATQTFEE471 pKa = 4.04VSLDD475 pKa = 3.47GMQEE479 pKa = 3.94VPAVDD484 pKa = 3.85TAATAAATVLLDD496 pKa = 3.6EE497 pKa = 5.44DD498 pKa = 4.66DD499 pKa = 4.66RR500 pKa = 11.84LFSVTVDD507 pKa = 3.21TSAVDD512 pKa = 3.39NVTGVHH518 pKa = 4.92VHH520 pKa = 7.22DD521 pKa = 5.15GNIGRR526 pKa = 11.84NGPVAFPLQASGDD539 pKa = 3.8DD540 pKa = 4.3TYY542 pKa = 11.62TLPATNLLDD551 pKa = 4.22PMIEE555 pKa = 3.97ALKK558 pKa = 10.59SGEE561 pKa = 3.88WYY563 pKa = 10.22INVHH567 pKa = 5.27TEE569 pKa = 3.88ANPSGEE575 pKa = 4.01VRR577 pKa = 11.84GQIVPEE583 pKa = 4.05TMAVVTFPLSGSQEE597 pKa = 4.35VPSVTTDD604 pKa = 2.63AMGYY608 pKa = 10.59GYY610 pKa = 9.02ATLDD614 pKa = 3.55TTTLAVDD621 pKa = 4.09LVAVTMGVEE630 pKa = 4.34DD631 pKa = 3.3ATMAHH636 pKa = 6.57IHH638 pKa = 5.92TGYY641 pKa = 10.6AGEE644 pKa = 4.34NGPVYY649 pKa = 9.8VTLEE653 pKa = 3.81QSMDD657 pKa = 3.82DD658 pKa = 3.59ANVWMTPAGAMIDD671 pKa = 3.54ATNAARR677 pKa = 11.84LVEE680 pKa = 4.37GGHH683 pKa = 5.74YY684 pKa = 10.93VNIHH688 pKa = 5.45TPANPSGEE696 pKa = 4.24LRR698 pKa = 11.84GQITPDD704 pKa = 3.38NIEE707 pKa = 4.21VYY709 pKa = 10.57GVTATGDD716 pKa = 3.59QEE718 pKa = 4.69VPAVDD723 pKa = 3.78TNAMGEE729 pKa = 4.25GAITLNTSTMTIKK742 pKa = 10.87AIINVMDD749 pKa = 4.07ISPNAAHH756 pKa = 7.0IHH758 pKa = 5.03QGAVGEE764 pKa = 4.21NGGVVIGLTNPDD776 pKa = 3.33AGLWTLDD783 pKa = 3.25ATLTEE788 pKa = 4.16AQMALMQAEE797 pKa = 4.01GLYY800 pKa = 10.82TNFHH804 pKa = 6.36TDD806 pKa = 3.12AFPSGEE812 pKa = 3.71IRR814 pKa = 11.84GQITLGFEE822 pKa = 4.07
Molecular weight: 85.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S2E7F6|A0A2S2E7F6_9ALTE OMP_b-brl domain-containing protein OS=Saliniradius amylolyticus OX=2183582 GN=HMF8227_02709 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIKK11 pKa = 10.11RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84SHH16 pKa = 6.18GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.42NGRR28 pKa = 11.84KK29 pKa = 8.55VLKK32 pKa = 10.18SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84ARR41 pKa = 11.84LAAA44 pKa = 4.44
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIKK11 pKa = 10.11RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84SHH16 pKa = 6.18GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.42NGRR28 pKa = 11.84KK29 pKa = 8.55VLKK32 pKa = 10.18SRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84ARR41 pKa = 11.84LAAA44 pKa = 4.44
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
972597 |
30 |
1835 |
330.3 |
36.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.898 ± 0.046 | 0.945 ± 0.017 |
5.918 ± 0.039 | 6.405 ± 0.046 |
3.831 ± 0.03 | 7.162 ± 0.038 |
2.397 ± 0.021 | 5.357 ± 0.032 |
4.535 ± 0.039 | 10.685 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.567 ± 0.022 | 3.723 ± 0.027 |
4.242 ± 0.03 | 5.368 ± 0.043 |
5.376 ± 0.036 | 6.203 ± 0.034 |
5.038 ± 0.03 | 6.946 ± 0.039 |
1.34 ± 0.019 | 3.065 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
