
Black grass cryptic virus 2
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
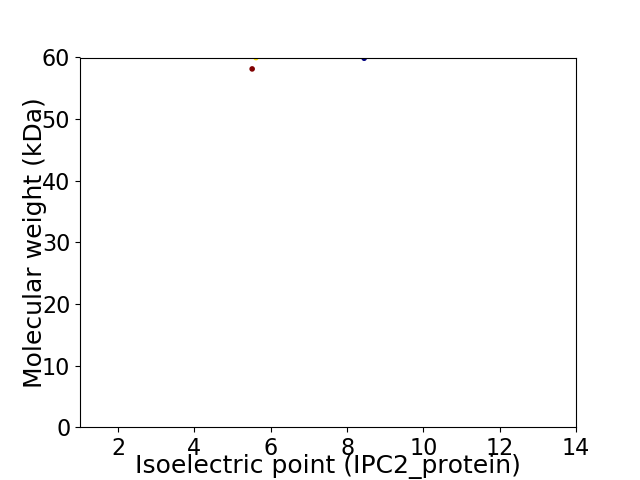
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D6DU17|A0A0D6DU17_9VIRU RNA dependent RNA polymerase OS=Black grass cryptic virus 2 OX=1587517 GN=RdRp PE=4 SV=1
MM1 pKa = 6.87STSEE5 pKa = 4.01GNLRR9 pKa = 11.84NDD11 pKa = 3.87PNRR14 pKa = 11.84IPEE17 pKa = 5.4PIPMDD22 pKa = 3.68IDD24 pKa = 3.7QLGHH28 pKa = 6.01SATDD32 pKa = 3.5VFDD35 pKa = 3.86PTHH38 pKa = 6.92FDD40 pKa = 3.59PEE42 pKa = 4.7GVKK45 pKa = 10.39PLVQDD50 pKa = 3.6PVAPKK55 pKa = 10.38KK56 pKa = 10.23RR57 pKa = 11.84GKK59 pKa = 6.55THH61 pKa = 6.59PRR63 pKa = 11.84SLDD66 pKa = 3.29KK67 pKa = 10.98QVVEE71 pKa = 4.45NSTPVNDD78 pKa = 3.92LRR80 pKa = 11.84RR81 pKa = 11.84VSASLAPSEE90 pKa = 5.41SRR92 pKa = 11.84FPDD95 pKa = 3.34PNMFIPNFVILFHH108 pKa = 7.03NLATMNNKK116 pKa = 8.62VCTMRR121 pKa = 11.84PFGEE125 pKa = 4.31SGTVNNWIPQVSNIVFAVLAIVQVLRR151 pKa = 11.84AQQLAGLLTPSQNRR165 pKa = 11.84FLKK168 pKa = 10.77SFIGKK173 pKa = 8.83HH174 pKa = 4.84PLEE177 pKa = 4.63TIIIPGPLLPFFRR190 pKa = 11.84SISSANPGFGNFGDD204 pKa = 4.12VTPVIPPFTQNEE216 pKa = 3.85EE217 pKa = 3.64RR218 pKa = 11.84LYY220 pKa = 10.78NIDD223 pKa = 4.27YY224 pKa = 10.35IPPSKK229 pKa = 10.53EE230 pKa = 3.64HH231 pKa = 5.64VFQGITMMLPNIPLMMDD248 pKa = 3.08QLVALVKK255 pKa = 10.55HH256 pKa = 5.82VLTLEE261 pKa = 3.74NNIPKK266 pKa = 10.09KK267 pKa = 10.82YY268 pKa = 9.51STFTQLQTIFGMNKK282 pKa = 9.56PEE284 pKa = 4.01EE285 pKa = 3.95QAPRR289 pKa = 11.84HH290 pKa = 5.19SAFVRR295 pKa = 11.84LQKK298 pKa = 10.91SIGFWQRR305 pKa = 11.84PTASDD310 pKa = 3.51GLIQRR315 pKa = 11.84FANYY319 pKa = 9.51IDD321 pKa = 4.4DD322 pKa = 4.44ACPDD326 pKa = 3.98FPAPPKK332 pKa = 10.45YY333 pKa = 10.59VSTHH337 pKa = 6.48AGTPADD343 pKa = 3.98PTQDD347 pKa = 3.4PPIPEE352 pKa = 4.51VPATWTALDD361 pKa = 4.39DD362 pKa = 4.27FAQFMCMEE370 pKa = 4.3EE371 pKa = 4.03TSVWFSKK378 pKa = 10.37FLEE381 pKa = 4.68GMHH384 pKa = 6.17VFNKK388 pKa = 8.25QWKK391 pKa = 8.94DD392 pKa = 3.36NEE394 pKa = 4.26RR395 pKa = 11.84LSNIHH400 pKa = 5.63PTKK403 pKa = 10.47SAAGLITFYY412 pKa = 10.74DD413 pKa = 3.68AEE415 pKa = 4.35YY416 pKa = 9.28TAIPTEE422 pKa = 4.04SQFRR426 pKa = 11.84DD427 pKa = 3.08RR428 pKa = 11.84SFNNATHH435 pKa = 5.74QLVMNTRR442 pKa = 11.84ATTACPHH449 pKa = 7.31IIPDD453 pKa = 4.11DD454 pKa = 5.11AIDD457 pKa = 4.08AQSSQLNAACARR469 pKa = 11.84VFNNAMTIGAPDD481 pKa = 3.49VTRR484 pKa = 11.84FGYY487 pKa = 10.25YY488 pKa = 8.24WVAKK492 pKa = 7.73PTTFQTNNGNLPDD505 pKa = 3.99MFPNVISSPDD515 pKa = 3.46YY516 pKa = 10.67FADD519 pKa = 3.41AA520 pKa = 4.46
MM1 pKa = 6.87STSEE5 pKa = 4.01GNLRR9 pKa = 11.84NDD11 pKa = 3.87PNRR14 pKa = 11.84IPEE17 pKa = 5.4PIPMDD22 pKa = 3.68IDD24 pKa = 3.7QLGHH28 pKa = 6.01SATDD32 pKa = 3.5VFDD35 pKa = 3.86PTHH38 pKa = 6.92FDD40 pKa = 3.59PEE42 pKa = 4.7GVKK45 pKa = 10.39PLVQDD50 pKa = 3.6PVAPKK55 pKa = 10.38KK56 pKa = 10.23RR57 pKa = 11.84GKK59 pKa = 6.55THH61 pKa = 6.59PRR63 pKa = 11.84SLDD66 pKa = 3.29KK67 pKa = 10.98QVVEE71 pKa = 4.45NSTPVNDD78 pKa = 3.92LRR80 pKa = 11.84RR81 pKa = 11.84VSASLAPSEE90 pKa = 5.41SRR92 pKa = 11.84FPDD95 pKa = 3.34PNMFIPNFVILFHH108 pKa = 7.03NLATMNNKK116 pKa = 8.62VCTMRR121 pKa = 11.84PFGEE125 pKa = 4.31SGTVNNWIPQVSNIVFAVLAIVQVLRR151 pKa = 11.84AQQLAGLLTPSQNRR165 pKa = 11.84FLKK168 pKa = 10.77SFIGKK173 pKa = 8.83HH174 pKa = 4.84PLEE177 pKa = 4.63TIIIPGPLLPFFRR190 pKa = 11.84SISSANPGFGNFGDD204 pKa = 4.12VTPVIPPFTQNEE216 pKa = 3.85EE217 pKa = 3.64RR218 pKa = 11.84LYY220 pKa = 10.78NIDD223 pKa = 4.27YY224 pKa = 10.35IPPSKK229 pKa = 10.53EE230 pKa = 3.64HH231 pKa = 5.64VFQGITMMLPNIPLMMDD248 pKa = 3.08QLVALVKK255 pKa = 10.55HH256 pKa = 5.82VLTLEE261 pKa = 3.74NNIPKK266 pKa = 10.09KK267 pKa = 10.82YY268 pKa = 9.51STFTQLQTIFGMNKK282 pKa = 9.56PEE284 pKa = 4.01EE285 pKa = 3.95QAPRR289 pKa = 11.84HH290 pKa = 5.19SAFVRR295 pKa = 11.84LQKK298 pKa = 10.91SIGFWQRR305 pKa = 11.84PTASDD310 pKa = 3.51GLIQRR315 pKa = 11.84FANYY319 pKa = 9.51IDD321 pKa = 4.4DD322 pKa = 4.44ACPDD326 pKa = 3.98FPAPPKK332 pKa = 10.45YY333 pKa = 10.59VSTHH337 pKa = 6.48AGTPADD343 pKa = 3.98PTQDD347 pKa = 3.4PPIPEE352 pKa = 4.51VPATWTALDD361 pKa = 4.39DD362 pKa = 4.27FAQFMCMEE370 pKa = 4.3EE371 pKa = 4.03TSVWFSKK378 pKa = 10.37FLEE381 pKa = 4.68GMHH384 pKa = 6.17VFNKK388 pKa = 8.25QWKK391 pKa = 8.94DD392 pKa = 3.36NEE394 pKa = 4.26RR395 pKa = 11.84LSNIHH400 pKa = 5.63PTKK403 pKa = 10.47SAAGLITFYY412 pKa = 10.74DD413 pKa = 3.68AEE415 pKa = 4.35YY416 pKa = 9.28TAIPTEE422 pKa = 4.04SQFRR426 pKa = 11.84DD427 pKa = 3.08RR428 pKa = 11.84SFNNATHH435 pKa = 5.74QLVMNTRR442 pKa = 11.84ATTACPHH449 pKa = 7.31IIPDD453 pKa = 4.11DD454 pKa = 5.11AIDD457 pKa = 4.08AQSSQLNAACARR469 pKa = 11.84VFNNAMTIGAPDD481 pKa = 3.49VTRR484 pKa = 11.84FGYY487 pKa = 10.25YY488 pKa = 8.24WVAKK492 pKa = 7.73PTTFQTNNGNLPDD505 pKa = 3.99MFPNVISSPDD515 pKa = 3.46YY516 pKa = 10.67FADD519 pKa = 3.41AA520 pKa = 4.46
Molecular weight: 58.12 kDa
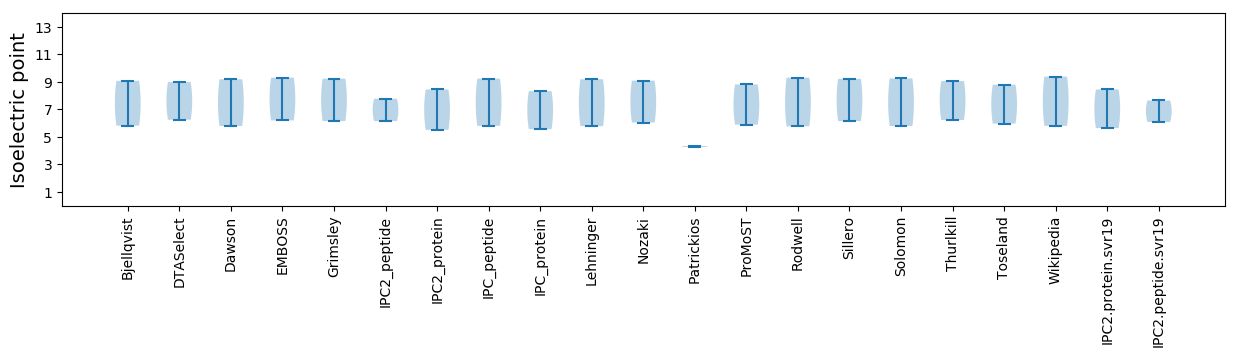
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D6DU17|A0A0D6DU17_9VIRU RNA dependent RNA polymerase OS=Black grass cryptic virus 2 OX=1587517 GN=RdRp PE=4 SV=1
MM1 pKa = 7.41TIEE4 pKa = 4.0AQYY7 pKa = 11.71EE8 pKa = 4.16DD9 pKa = 4.6MLKK12 pKa = 9.58MNQPPVHH19 pKa = 6.48IVQDD23 pKa = 4.02EE24 pKa = 4.39NFTTAFNLTMKK35 pKa = 10.72AFAPTEE41 pKa = 3.89KK42 pKa = 10.35VRR44 pKa = 11.84IVHH47 pKa = 5.33YY48 pKa = 10.71VRR50 pKa = 11.84TKK52 pKa = 10.01HH53 pKa = 5.75YY54 pKa = 7.67PWPLSSSVEE63 pKa = 4.25RR64 pKa = 11.84PFSTDD69 pKa = 2.83PKK71 pKa = 8.5VIKK74 pKa = 10.01YY75 pKa = 8.87VKK77 pKa = 9.48QKK79 pKa = 10.45CINGEE84 pKa = 3.89IEE86 pKa = 3.91NEE88 pKa = 3.58RR89 pKa = 11.84MSFHH93 pKa = 5.98NTFDD97 pKa = 3.54YY98 pKa = 11.33VYY100 pKa = 10.14DD101 pKa = 3.62SSRR104 pKa = 11.84LVIHH108 pKa = 6.72NIKK111 pKa = 10.28NGKK114 pKa = 9.2CFTGDD119 pKa = 3.14LTKK122 pKa = 11.17DD123 pKa = 3.06HH124 pKa = 7.58LYY126 pKa = 10.29PIIAHH131 pKa = 6.13FRR133 pKa = 11.84PGLGKK138 pKa = 9.76PGSSAIKK145 pKa = 10.15NRR147 pKa = 11.84LVWGVSKK154 pKa = 10.73IFLIAEE160 pKa = 4.28CMFMYY165 pKa = 10.61PLFDD169 pKa = 3.52NYY171 pKa = 10.54LRR173 pKa = 11.84NGKK176 pKa = 7.62TPLLWKK182 pKa = 10.81YY183 pKa = 7.38EE184 pKa = 4.1TALGGWTKK192 pKa = 10.66LYY194 pKa = 11.24NEE196 pKa = 4.78IQPQIANTNATIITADD212 pKa = 2.76WSEE215 pKa = 3.87FDD217 pKa = 3.16RR218 pKa = 11.84RR219 pKa = 11.84VPFEE223 pKa = 4.77LIEE226 pKa = 4.04MVINASLEE234 pKa = 4.22YY235 pKa = 10.64YY236 pKa = 10.69DD237 pKa = 4.97LGHH240 pKa = 6.41YY241 pKa = 10.22APDD244 pKa = 3.63YY245 pKa = 10.7LYY247 pKa = 9.25HH248 pKa = 6.44TEE250 pKa = 4.23SNCKK254 pKa = 10.18YY255 pKa = 9.12EE256 pKa = 4.16IPRR259 pKa = 11.84ILDD262 pKa = 3.37LFKK265 pKa = 8.74WTIYY269 pKa = 9.55ATLSSPLLMPDD280 pKa = 2.51GRR282 pKa = 11.84VWMRR286 pKa = 11.84TRR288 pKa = 11.84NGVPSGMFRR297 pKa = 11.84TQWLDD302 pKa = 3.48SIINGIMITTILLDD316 pKa = 3.81AGFKK320 pKa = 8.97VTEE323 pKa = 4.04KK324 pKa = 10.88FILKK328 pKa = 10.26VLGDD332 pKa = 4.13DD333 pKa = 3.62SLTVLFHH340 pKa = 6.79FVPQAKK346 pKa = 10.36HH347 pKa = 6.1EE348 pKa = 4.17DD349 pKa = 3.66LKK351 pKa = 11.68SFLAEE356 pKa = 3.79KK357 pKa = 10.69ALLRR361 pKa = 11.84FNAKK365 pKa = 10.23LSLEE369 pKa = 4.01KK370 pKa = 10.54TEE372 pKa = 4.83ISNSLYY378 pKa = 10.32GVEE381 pKa = 3.85LLGYY385 pKa = 9.86RR386 pKa = 11.84NRR388 pKa = 11.84NGAAYY393 pKa = 9.64RR394 pKa = 11.84DD395 pKa = 3.92HH396 pKa = 7.53RR397 pKa = 11.84KK398 pKa = 9.55LLAQLLFPEE407 pKa = 5.08SEE409 pKa = 4.22APTYY413 pKa = 10.72ASLMGRR419 pKa = 11.84CVGIAYY425 pKa = 10.17ADD427 pKa = 3.73LGRR430 pKa = 11.84SKK432 pKa = 11.06LLYY435 pKa = 9.67RR436 pKa = 11.84VCKK439 pKa = 10.4KK440 pKa = 10.19IFEE443 pKa = 4.44RR444 pKa = 11.84LSAAGYY450 pKa = 10.2RR451 pKa = 11.84PDD453 pKa = 2.83SRR455 pKa = 11.84QIHH458 pKa = 6.62RR459 pKa = 11.84YY460 pKa = 8.97LSGSKK465 pKa = 9.96FVEE468 pKa = 3.91TRR470 pKa = 11.84MEE472 pKa = 3.6IQINKK477 pKa = 9.08FPSTLEE483 pKa = 3.54IQKK486 pKa = 9.81WSRR489 pKa = 11.84TPYY492 pKa = 10.77NRR494 pKa = 11.84TKK496 pKa = 10.88SDD498 pKa = 3.54NDD500 pKa = 4.01HH501 pKa = 6.31YY502 pKa = 10.56WPPEE506 pKa = 3.66IFWPHH511 pKa = 4.44FTGPP515 pKa = 3.39
MM1 pKa = 7.41TIEE4 pKa = 4.0AQYY7 pKa = 11.71EE8 pKa = 4.16DD9 pKa = 4.6MLKK12 pKa = 9.58MNQPPVHH19 pKa = 6.48IVQDD23 pKa = 4.02EE24 pKa = 4.39NFTTAFNLTMKK35 pKa = 10.72AFAPTEE41 pKa = 3.89KK42 pKa = 10.35VRR44 pKa = 11.84IVHH47 pKa = 5.33YY48 pKa = 10.71VRR50 pKa = 11.84TKK52 pKa = 10.01HH53 pKa = 5.75YY54 pKa = 7.67PWPLSSSVEE63 pKa = 4.25RR64 pKa = 11.84PFSTDD69 pKa = 2.83PKK71 pKa = 8.5VIKK74 pKa = 10.01YY75 pKa = 8.87VKK77 pKa = 9.48QKK79 pKa = 10.45CINGEE84 pKa = 3.89IEE86 pKa = 3.91NEE88 pKa = 3.58RR89 pKa = 11.84MSFHH93 pKa = 5.98NTFDD97 pKa = 3.54YY98 pKa = 11.33VYY100 pKa = 10.14DD101 pKa = 3.62SSRR104 pKa = 11.84LVIHH108 pKa = 6.72NIKK111 pKa = 10.28NGKK114 pKa = 9.2CFTGDD119 pKa = 3.14LTKK122 pKa = 11.17DD123 pKa = 3.06HH124 pKa = 7.58LYY126 pKa = 10.29PIIAHH131 pKa = 6.13FRR133 pKa = 11.84PGLGKK138 pKa = 9.76PGSSAIKK145 pKa = 10.15NRR147 pKa = 11.84LVWGVSKK154 pKa = 10.73IFLIAEE160 pKa = 4.28CMFMYY165 pKa = 10.61PLFDD169 pKa = 3.52NYY171 pKa = 10.54LRR173 pKa = 11.84NGKK176 pKa = 7.62TPLLWKK182 pKa = 10.81YY183 pKa = 7.38EE184 pKa = 4.1TALGGWTKK192 pKa = 10.66LYY194 pKa = 11.24NEE196 pKa = 4.78IQPQIANTNATIITADD212 pKa = 2.76WSEE215 pKa = 3.87FDD217 pKa = 3.16RR218 pKa = 11.84RR219 pKa = 11.84VPFEE223 pKa = 4.77LIEE226 pKa = 4.04MVINASLEE234 pKa = 4.22YY235 pKa = 10.64YY236 pKa = 10.69DD237 pKa = 4.97LGHH240 pKa = 6.41YY241 pKa = 10.22APDD244 pKa = 3.63YY245 pKa = 10.7LYY247 pKa = 9.25HH248 pKa = 6.44TEE250 pKa = 4.23SNCKK254 pKa = 10.18YY255 pKa = 9.12EE256 pKa = 4.16IPRR259 pKa = 11.84ILDD262 pKa = 3.37LFKK265 pKa = 8.74WTIYY269 pKa = 9.55ATLSSPLLMPDD280 pKa = 2.51GRR282 pKa = 11.84VWMRR286 pKa = 11.84TRR288 pKa = 11.84NGVPSGMFRR297 pKa = 11.84TQWLDD302 pKa = 3.48SIINGIMITTILLDD316 pKa = 3.81AGFKK320 pKa = 8.97VTEE323 pKa = 4.04KK324 pKa = 10.88FILKK328 pKa = 10.26VLGDD332 pKa = 4.13DD333 pKa = 3.62SLTVLFHH340 pKa = 6.79FVPQAKK346 pKa = 10.36HH347 pKa = 6.1EE348 pKa = 4.17DD349 pKa = 3.66LKK351 pKa = 11.68SFLAEE356 pKa = 3.79KK357 pKa = 10.69ALLRR361 pKa = 11.84FNAKK365 pKa = 10.23LSLEE369 pKa = 4.01KK370 pKa = 10.54TEE372 pKa = 4.83ISNSLYY378 pKa = 10.32GVEE381 pKa = 3.85LLGYY385 pKa = 9.86RR386 pKa = 11.84NRR388 pKa = 11.84NGAAYY393 pKa = 9.64RR394 pKa = 11.84DD395 pKa = 3.92HH396 pKa = 7.53RR397 pKa = 11.84KK398 pKa = 9.55LLAQLLFPEE407 pKa = 5.08SEE409 pKa = 4.22APTYY413 pKa = 10.72ASLMGRR419 pKa = 11.84CVGIAYY425 pKa = 10.17ADD427 pKa = 3.73LGRR430 pKa = 11.84SKK432 pKa = 11.06LLYY435 pKa = 9.67RR436 pKa = 11.84VCKK439 pKa = 10.4KK440 pKa = 10.19IFEE443 pKa = 4.44RR444 pKa = 11.84LSAAGYY450 pKa = 10.2RR451 pKa = 11.84PDD453 pKa = 2.83SRR455 pKa = 11.84QIHH458 pKa = 6.62RR459 pKa = 11.84YY460 pKa = 8.97LSGSKK465 pKa = 9.96FVEE468 pKa = 3.91TRR470 pKa = 11.84MEE472 pKa = 3.6IQINKK477 pKa = 9.08FPSTLEE483 pKa = 3.54IQKK486 pKa = 9.81WSRR489 pKa = 11.84TPYY492 pKa = 10.77NRR494 pKa = 11.84TKK496 pKa = 10.88SDD498 pKa = 3.54NDD500 pKa = 4.01HH501 pKa = 6.31YY502 pKa = 10.56WPPEE506 pKa = 3.66IFWPHH511 pKa = 4.44FTGPP515 pKa = 3.39
Molecular weight: 59.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1035 |
515 |
520 |
517.5 |
58.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.473 ± 0.679 | 1.063 ± 0.067 |
5.314 ± 0.428 | 4.831 ± 0.652 |
5.99 ± 0.49 | 4.638 ± 0.269 |
2.705 ± 0.136 | 6.57 ± 0.275 |
5.314 ± 0.971 | 8.213 ± 1.108 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.899 ± 0.118 | 5.894 ± 0.681 |
7.923 ± 1.502 | 3.575 ± 0.816 |
5.024 ± 0.525 | 6.184 ± 0.02 |
6.667 ± 0.297 | 5.411 ± 0.492 |
1.643 ± 0.323 | 3.671 ± 1.157 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
