
Bizionia argentinensis JUB59
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Bizionia; Bizionia argentinensis
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
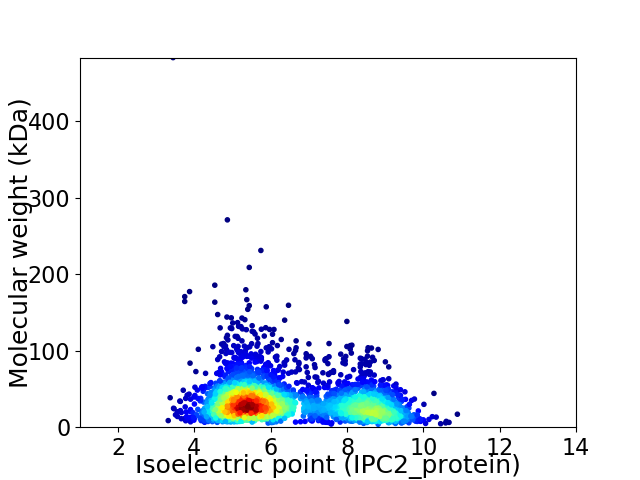
Virtual 2D-PAGE plot for 2926 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G2EGL8|G2EGL8_9FLAO tRNA (Guanine-N1)-methyltransferase OS=Bizionia argentinensis JUB59 OX=1046627 GN=BZARG_2276 PE=4 SV=1
MM1 pKa = 7.67NLRR4 pKa = 11.84KK5 pKa = 9.58ILVLLLAVSSVITACKK21 pKa = 10.01KK22 pKa = 10.77DD23 pKa = 4.24DD24 pKa = 4.78DD25 pKa = 5.05DD26 pKa = 6.95GPMTVEE32 pKa = 4.13LRR34 pKa = 11.84DD35 pKa = 3.46AAEE38 pKa = 4.49VYY40 pKa = 10.74AEE42 pKa = 4.66DD43 pKa = 4.58MIDD46 pKa = 3.22IQDD49 pKa = 3.63YY50 pKa = 11.37LEE52 pKa = 4.03THH54 pKa = 7.05FYY56 pKa = 10.78NYY58 pKa = 10.62EE59 pKa = 3.91EE60 pKa = 4.24FQLNSAYY67 pKa = 10.98SLANDD72 pKa = 3.25NFKK75 pKa = 10.7IVFDD79 pKa = 4.88TIAGTNANKK88 pKa = 9.61TPLIDD93 pKa = 3.67MVDD96 pKa = 3.68SKK98 pKa = 11.27IVTQGGIDD106 pKa = 3.38YY107 pKa = 8.64TFYY110 pKa = 10.86YY111 pKa = 10.74LNVRR115 pKa = 11.84EE116 pKa = 4.11GLGNEE121 pKa = 3.64IHH123 pKa = 7.09RR124 pKa = 11.84SDD126 pKa = 4.16EE127 pKa = 3.66ATLIYY132 pKa = 9.98EE133 pKa = 4.86GSSIPDD139 pKa = 3.06NYY141 pKa = 10.68VFDD144 pKa = 4.04SSVNEE149 pKa = 4.11SQFNLMAVGATSGVIRR165 pKa = 11.84GFMEE169 pKa = 4.04GVIEE173 pKa = 4.93FKK175 pKa = 10.69TSDD178 pKa = 3.55SYY180 pKa = 11.89DD181 pKa = 3.32INGDD185 pKa = 3.39GTVAYY190 pKa = 9.36HH191 pKa = 5.19NHH193 pKa = 6.88GIGAAFIPSGIGYY206 pKa = 9.17FNQPLAGVPSYY217 pKa = 10.95TPLIFKK223 pKa = 10.4FSLYY227 pKa = 10.45KK228 pKa = 9.86RR229 pKa = 11.84TILDD233 pKa = 3.63HH234 pKa = 7.42DD235 pKa = 4.78LDD237 pKa = 5.4GIPSYY242 pKa = 11.42LEE244 pKa = 4.18DD245 pKa = 5.08LNNNDD250 pKa = 5.63DD251 pKa = 5.53LFDD254 pKa = 6.13DD255 pKa = 4.95DD256 pKa = 4.68TDD258 pKa = 4.31GDD260 pKa = 3.86QTPNFLDD267 pKa = 3.97NDD269 pKa = 4.38DD270 pKa = 5.67DD271 pKa = 5.93GDD273 pKa = 4.2GYY275 pKa = 9.49LTKK278 pKa = 10.85DD279 pKa = 3.11EE280 pKa = 4.99LDD282 pKa = 3.4YY283 pKa = 11.66GEE285 pKa = 5.07YY286 pKa = 10.17VIQVGDD292 pKa = 3.75SDD294 pKa = 3.86PTFAANEE301 pKa = 4.21FEE303 pKa = 4.4SSRR306 pKa = 11.84VEE308 pKa = 3.78NNGQITIKK316 pKa = 9.95TVILRR321 pKa = 11.84DD322 pKa = 3.64TNGNGTPDD330 pKa = 3.76YY331 pKa = 10.79LDD333 pKa = 3.51EE334 pKa = 4.44TSIPDD339 pKa = 3.24
MM1 pKa = 7.67NLRR4 pKa = 11.84KK5 pKa = 9.58ILVLLLAVSSVITACKK21 pKa = 10.01KK22 pKa = 10.77DD23 pKa = 4.24DD24 pKa = 4.78DD25 pKa = 5.05DD26 pKa = 6.95GPMTVEE32 pKa = 4.13LRR34 pKa = 11.84DD35 pKa = 3.46AAEE38 pKa = 4.49VYY40 pKa = 10.74AEE42 pKa = 4.66DD43 pKa = 4.58MIDD46 pKa = 3.22IQDD49 pKa = 3.63YY50 pKa = 11.37LEE52 pKa = 4.03THH54 pKa = 7.05FYY56 pKa = 10.78NYY58 pKa = 10.62EE59 pKa = 3.91EE60 pKa = 4.24FQLNSAYY67 pKa = 10.98SLANDD72 pKa = 3.25NFKK75 pKa = 10.7IVFDD79 pKa = 4.88TIAGTNANKK88 pKa = 9.61TPLIDD93 pKa = 3.67MVDD96 pKa = 3.68SKK98 pKa = 11.27IVTQGGIDD106 pKa = 3.38YY107 pKa = 8.64TFYY110 pKa = 10.86YY111 pKa = 10.74LNVRR115 pKa = 11.84EE116 pKa = 4.11GLGNEE121 pKa = 3.64IHH123 pKa = 7.09RR124 pKa = 11.84SDD126 pKa = 4.16EE127 pKa = 3.66ATLIYY132 pKa = 9.98EE133 pKa = 4.86GSSIPDD139 pKa = 3.06NYY141 pKa = 10.68VFDD144 pKa = 4.04SSVNEE149 pKa = 4.11SQFNLMAVGATSGVIRR165 pKa = 11.84GFMEE169 pKa = 4.04GVIEE173 pKa = 4.93FKK175 pKa = 10.69TSDD178 pKa = 3.55SYY180 pKa = 11.89DD181 pKa = 3.32INGDD185 pKa = 3.39GTVAYY190 pKa = 9.36HH191 pKa = 5.19NHH193 pKa = 6.88GIGAAFIPSGIGYY206 pKa = 9.17FNQPLAGVPSYY217 pKa = 10.95TPLIFKK223 pKa = 10.4FSLYY227 pKa = 10.45KK228 pKa = 9.86RR229 pKa = 11.84TILDD233 pKa = 3.63HH234 pKa = 7.42DD235 pKa = 4.78LDD237 pKa = 5.4GIPSYY242 pKa = 11.42LEE244 pKa = 4.18DD245 pKa = 5.08LNNNDD250 pKa = 5.63DD251 pKa = 5.53LFDD254 pKa = 6.13DD255 pKa = 4.95DD256 pKa = 4.68TDD258 pKa = 4.31GDD260 pKa = 3.86QTPNFLDD267 pKa = 3.97NDD269 pKa = 4.38DD270 pKa = 5.67DD271 pKa = 5.93GDD273 pKa = 4.2GYY275 pKa = 9.49LTKK278 pKa = 10.85DD279 pKa = 3.11EE280 pKa = 4.99LDD282 pKa = 3.4YY283 pKa = 11.66GEE285 pKa = 5.07YY286 pKa = 10.17VIQVGDD292 pKa = 3.75SDD294 pKa = 3.86PTFAANEE301 pKa = 4.21FEE303 pKa = 4.4SSRR306 pKa = 11.84VEE308 pKa = 3.78NNGQITIKK316 pKa = 9.95TVILRR321 pKa = 11.84DD322 pKa = 3.64TNGNGTPDD330 pKa = 3.76YY331 pKa = 10.79LDD333 pKa = 3.51EE334 pKa = 4.44TSIPDD339 pKa = 3.24
Molecular weight: 37.63 kDa
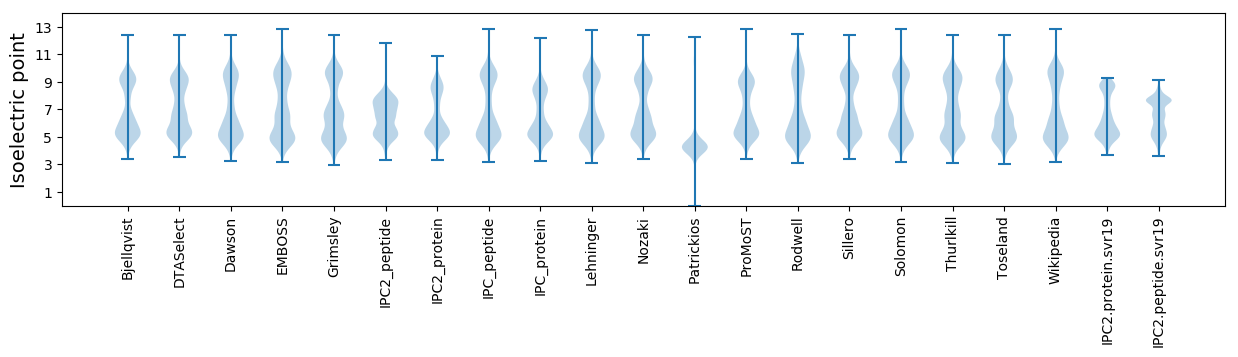
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G2EG69|G2EG69_9FLAO Tetratricopeptide repeat protein OS=Bizionia argentinensis JUB59 OX=1046627 GN=BZARG_1903 PE=4 SV=1
MM1 pKa = 7.7NIFLKK6 pKa = 10.63PLLYY10 pKa = 9.41FQKK13 pKa = 10.56HH14 pKa = 5.12GFHH17 pKa = 5.9VCQRR21 pKa = 11.84IADD24 pKa = 4.0RR25 pKa = 11.84MGIRR29 pKa = 11.84AKK31 pKa = 9.99VVRR34 pKa = 11.84TSFMYY39 pKa = 9.36LTFVTLGFGFAVYY52 pKa = 10.09LFIAFWMRR60 pKa = 11.84IKK62 pKa = 10.93DD63 pKa = 3.65LLFTKK68 pKa = 10.28RR69 pKa = 11.84SSVFDD74 pKa = 3.68LL75 pKa = 4.35
MM1 pKa = 7.7NIFLKK6 pKa = 10.63PLLYY10 pKa = 9.41FQKK13 pKa = 10.56HH14 pKa = 5.12GFHH17 pKa = 5.9VCQRR21 pKa = 11.84IADD24 pKa = 4.0RR25 pKa = 11.84MGIRR29 pKa = 11.84AKK31 pKa = 9.99VVRR34 pKa = 11.84TSFMYY39 pKa = 9.36LTFVTLGFGFAVYY52 pKa = 10.09LFIAFWMRR60 pKa = 11.84IKK62 pKa = 10.93DD63 pKa = 3.65LLFTKK68 pKa = 10.28RR69 pKa = 11.84SSVFDD74 pKa = 3.68LL75 pKa = 4.35
Molecular weight: 8.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
948185 |
34 |
4606 |
324.1 |
36.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.454 ± 0.037 | 0.714 ± 0.015 |
5.649 ± 0.031 | 6.523 ± 0.044 |
5.322 ± 0.04 | 6.109 ± 0.047 |
1.83 ± 0.018 | 8.264 ± 0.047 |
7.796 ± 0.057 | 9.378 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.296 ± 0.021 | 6.23 ± 0.039 |
3.253 ± 0.025 | 3.372 ± 0.027 |
3.32 ± 0.031 | 6.509 ± 0.031 |
5.837 ± 0.057 | 6.173 ± 0.036 |
0.95 ± 0.017 | 4.022 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
