
Tortoise microvirus 43
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
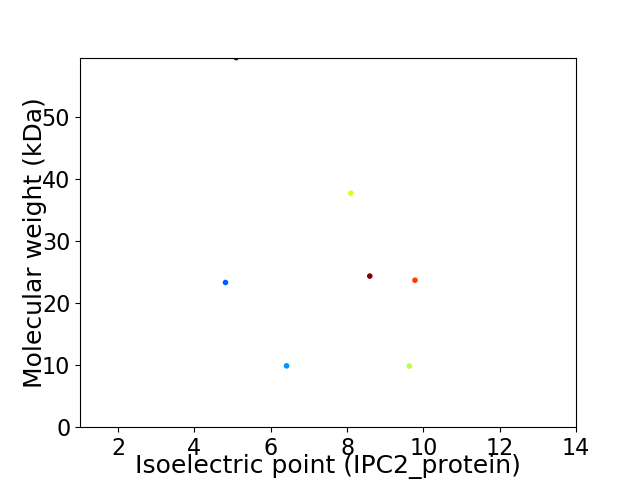
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
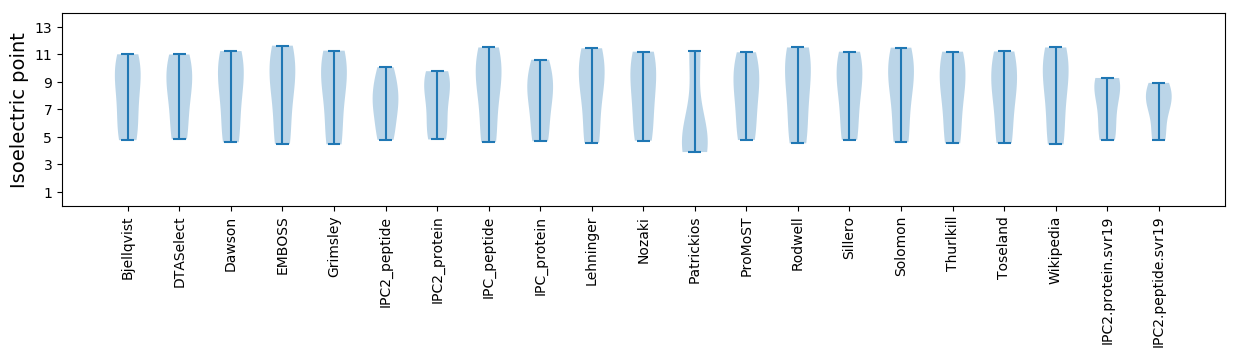
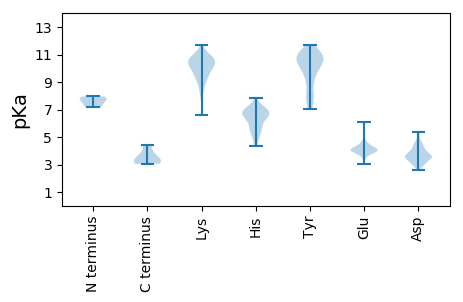
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W792|A0A4P8W792_9VIRU Replication initiation protein OS=Tortoise microvirus 43 OX=2583147 PE=4 SV=1
MM1 pKa = 7.95DD2 pKa = 4.2RR3 pKa = 11.84QRR5 pKa = 11.84EE6 pKa = 4.14EE7 pKa = 4.05TGGYY11 pKa = 7.66TYY13 pKa = 10.77SGSSSEE19 pKa = 3.98YY20 pKa = 9.54SAYY23 pKa = 10.01EE24 pKa = 4.01AYY26 pKa = 10.2QKK28 pKa = 11.4GLIGGGGNGGTGKK41 pKa = 8.23TVTAGPGVDD50 pKa = 3.72GAQTVPVGEE59 pKa = 4.46PVAEE63 pKa = 4.15VAPVTQQTPPAPVTGPGGPRR83 pKa = 11.84RR84 pKa = 11.84QSTLLSPLEE93 pKa = 3.92PMGPYY98 pKa = 9.97GPRR101 pKa = 11.84IQRR104 pKa = 11.84EE105 pKa = 4.68GTRR108 pKa = 11.84LTPKK112 pKa = 9.8MEE114 pKa = 4.1MSVQEE119 pKa = 4.2MLVDD123 pKa = 4.55GYY125 pKa = 10.91RR126 pKa = 11.84INANPRR132 pKa = 11.84TSNAEE137 pKa = 3.57EE138 pKa = 3.99WEE140 pKa = 3.87IRR142 pKa = 11.84YY143 pKa = 9.65AEE145 pKa = 4.4PGEE148 pKa = 4.15WVGGIVVMASDD159 pKa = 3.07AWYY162 pKa = 7.99NTKK165 pKa = 10.07QALGDD170 pKa = 3.73LSYY173 pKa = 11.19KK174 pKa = 10.83ANAANNSWAGATHH187 pKa = 7.07AGDD190 pKa = 5.36AFGDD194 pKa = 3.69WFDD197 pKa = 4.58SRR199 pKa = 11.84LQSVSPKK206 pKa = 10.44AGEE209 pKa = 4.07LVQEE213 pKa = 4.21PVDD216 pKa = 3.31WGRR219 pKa = 3.47
MM1 pKa = 7.95DD2 pKa = 4.2RR3 pKa = 11.84QRR5 pKa = 11.84EE6 pKa = 4.14EE7 pKa = 4.05TGGYY11 pKa = 7.66TYY13 pKa = 10.77SGSSSEE19 pKa = 3.98YY20 pKa = 9.54SAYY23 pKa = 10.01EE24 pKa = 4.01AYY26 pKa = 10.2QKK28 pKa = 11.4GLIGGGGNGGTGKK41 pKa = 8.23TVTAGPGVDD50 pKa = 3.72GAQTVPVGEE59 pKa = 4.46PVAEE63 pKa = 4.15VAPVTQQTPPAPVTGPGGPRR83 pKa = 11.84RR84 pKa = 11.84QSTLLSPLEE93 pKa = 3.92PMGPYY98 pKa = 9.97GPRR101 pKa = 11.84IQRR104 pKa = 11.84EE105 pKa = 4.68GTRR108 pKa = 11.84LTPKK112 pKa = 9.8MEE114 pKa = 4.1MSVQEE119 pKa = 4.2MLVDD123 pKa = 4.55GYY125 pKa = 10.91RR126 pKa = 11.84INANPRR132 pKa = 11.84TSNAEE137 pKa = 3.57EE138 pKa = 3.99WEE140 pKa = 3.87IRR142 pKa = 11.84YY143 pKa = 9.65AEE145 pKa = 4.4PGEE148 pKa = 4.15WVGGIVVMASDD159 pKa = 3.07AWYY162 pKa = 7.99NTKK165 pKa = 10.07QALGDD170 pKa = 3.73LSYY173 pKa = 11.19KK174 pKa = 10.83ANAANNSWAGATHH187 pKa = 7.07AGDD190 pKa = 5.36AFGDD194 pKa = 3.69WFDD197 pKa = 4.58SRR199 pKa = 11.84LQSVSPKK206 pKa = 10.44AGEE209 pKa = 4.07LVQEE213 pKa = 4.21PVDD216 pKa = 3.31WGRR219 pKa = 3.47
Molecular weight: 23.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W9S0|A0A4P8W9S0_9VIRU Major capsid protein OS=Tortoise microvirus 43 OX=2583147 PE=4 SV=1
MM1 pKa = 7.89ANRR4 pKa = 11.84TGNTSKK10 pKa = 10.95EE11 pKa = 4.07KK12 pKa = 10.89AEE14 pKa = 3.72RR15 pKa = 11.84AARR18 pKa = 11.84NTSSASSGSRR28 pKa = 11.84TSSSSRR34 pKa = 11.84SNSGTSKK41 pKa = 10.67ASSGSKK47 pKa = 10.21GVSTPRR53 pKa = 11.84TSSRR57 pKa = 11.84QSAPRR62 pKa = 11.84APDD65 pKa = 3.36PVSRR69 pKa = 11.84QSSRR73 pKa = 11.84ATARR77 pKa = 11.84ADD79 pKa = 3.61ASRR82 pKa = 11.84GDD84 pKa = 4.02HH85 pKa = 5.93GWNDD89 pKa = 2.96SWAGASTDD97 pKa = 3.45NGSIGSNSGSDD108 pKa = 3.77YY109 pKa = 11.33GWGNAKK115 pKa = 10.22KK116 pKa = 9.74KK117 pKa = 8.72VTGGTPTSPSPSLPSPTAPDD137 pKa = 3.42PVEE140 pKa = 3.99TRR142 pKa = 11.84KK143 pKa = 9.85SAKK146 pKa = 8.9ATNLRR151 pKa = 11.84IAPTRR156 pKa = 11.84EE157 pKa = 3.85TLPHH161 pKa = 6.75KK162 pKa = 10.01EE163 pKa = 3.72RR164 pKa = 11.84SPARR168 pKa = 11.84QEE170 pKa = 3.87ARR172 pKa = 11.84AKK174 pKa = 10.11RR175 pKa = 11.84STPIEE180 pKa = 3.98PAKK183 pKa = 10.28QSPSPKK189 pKa = 9.39VRR191 pKa = 11.84EE192 pKa = 3.89EE193 pKa = 4.02KK194 pKa = 10.79KK195 pKa = 10.32PVLHH199 pKa = 6.52CKK201 pKa = 9.52PRR203 pKa = 11.84PKK205 pKa = 10.4DD206 pKa = 3.6NKK208 pKa = 8.18PTGGGGGGGNFKK220 pKa = 10.72KK221 pKa = 10.68FIPWCGG227 pKa = 3.05
MM1 pKa = 7.89ANRR4 pKa = 11.84TGNTSKK10 pKa = 10.95EE11 pKa = 4.07KK12 pKa = 10.89AEE14 pKa = 3.72RR15 pKa = 11.84AARR18 pKa = 11.84NTSSASSGSRR28 pKa = 11.84TSSSSRR34 pKa = 11.84SNSGTSKK41 pKa = 10.67ASSGSKK47 pKa = 10.21GVSTPRR53 pKa = 11.84TSSRR57 pKa = 11.84QSAPRR62 pKa = 11.84APDD65 pKa = 3.36PVSRR69 pKa = 11.84QSSRR73 pKa = 11.84ATARR77 pKa = 11.84ADD79 pKa = 3.61ASRR82 pKa = 11.84GDD84 pKa = 4.02HH85 pKa = 5.93GWNDD89 pKa = 2.96SWAGASTDD97 pKa = 3.45NGSIGSNSGSDD108 pKa = 3.77YY109 pKa = 11.33GWGNAKK115 pKa = 10.22KK116 pKa = 9.74KK117 pKa = 8.72VTGGTPTSPSPSLPSPTAPDD137 pKa = 3.42PVEE140 pKa = 3.99TRR142 pKa = 11.84KK143 pKa = 9.85SAKK146 pKa = 8.9ATNLRR151 pKa = 11.84IAPTRR156 pKa = 11.84EE157 pKa = 3.85TLPHH161 pKa = 6.75KK162 pKa = 10.01EE163 pKa = 3.72RR164 pKa = 11.84SPARR168 pKa = 11.84QEE170 pKa = 3.87ARR172 pKa = 11.84AKK174 pKa = 10.11RR175 pKa = 11.84STPIEE180 pKa = 3.98PAKK183 pKa = 10.28QSPSPKK189 pKa = 9.39VRR191 pKa = 11.84EE192 pKa = 3.89EE193 pKa = 4.02KK194 pKa = 10.79KK195 pKa = 10.32PVLHH199 pKa = 6.52CKK201 pKa = 9.52PRR203 pKa = 11.84PKK205 pKa = 10.4DD206 pKa = 3.6NKK208 pKa = 8.18PTGGGGGGGNFKK220 pKa = 10.72KK221 pKa = 10.68FIPWCGG227 pKa = 3.05
Molecular weight: 23.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1693 |
89 |
539 |
241.9 |
26.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.451 ± 1.037 | 0.413 ± 0.19 |
5.08 ± 0.601 | 7.324 ± 0.746 |
2.953 ± 0.694 | 8.683 ± 1.145 |
2.304 ± 0.527 | 3.131 ± 0.418 |
5.67 ± 1.135 | 6.852 ± 1.227 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.481 ± 0.478 | 3.249 ± 0.745 |
7.147 ± 0.888 | 4.194 ± 0.736 |
7.915 ± 0.724 | 6.615 ± 1.991 |
5.316 ± 1.101 | 6.143 ± 0.894 |
1.949 ± 0.409 | 3.131 ± 0.787 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
