
Nonomuraea solani
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Streptosporangiaceae; Nonomuraea
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
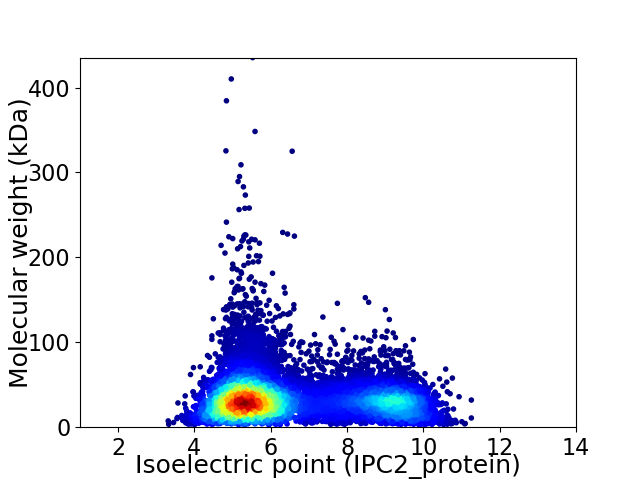
Virtual 2D-PAGE plot for 12105 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H6D4K4|A0A1H6D4K4_9ACTN Uncharacterized protein OS=Nonomuraea solani OX=1144553 GN=SAMN05444920_104720 PE=4 SV=1
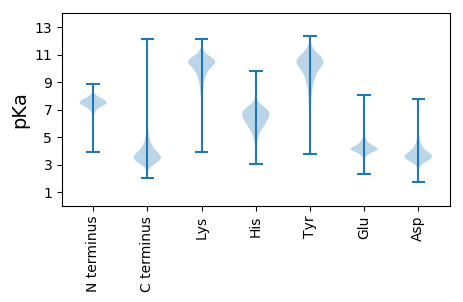
MM1 pKa = 7.17NPIVPTAKK9 pKa = 10.18RR10 pKa = 11.84GIGAAALAAGMVLASPITAGAAAAPPATYY39 pKa = 10.61ASVVGDD45 pKa = 3.68TLNVDD50 pKa = 3.92GNYY53 pKa = 10.01EE54 pKa = 4.41SNHH57 pKa = 4.6ITITPAAYY65 pKa = 9.91GRR67 pKa = 11.84VRR69 pKa = 11.84IADD72 pKa = 3.84ANNPTLPGGDD82 pKa = 4.48CIDD85 pKa = 3.65ITDD88 pKa = 4.32HH89 pKa = 6.57EE90 pKa = 5.07VEE92 pKa = 5.0CGLSGTPTGLDD103 pKa = 3.19VDD105 pKa = 4.05GHH107 pKa = 6.32EE108 pKa = 4.85GADD111 pKa = 3.69TIINLLPTSNKK122 pKa = 7.47TQARR126 pKa = 11.84LHH128 pKa = 6.36GGSGNDD134 pKa = 3.51VIYY137 pKa = 10.44GGPGVQWLNGGLSPDD152 pKa = 4.5DD153 pKa = 3.74LAANQGRR160 pKa = 11.84PDD162 pKa = 3.52TGNDD166 pKa = 3.25KK167 pKa = 11.24LFGGCAQEE175 pKa = 4.76CADD178 pKa = 5.29SGDD181 pKa = 4.04LLEE184 pKa = 5.95GSDD187 pKa = 4.59GNDD190 pKa = 3.36GLTGGPGNDD199 pKa = 4.29DD200 pKa = 3.43LRR202 pKa = 11.84GGIGVDD208 pKa = 3.85TYY210 pKa = 11.7AGGSGRR216 pKa = 11.84DD217 pKa = 3.45VVSYY221 pKa = 10.78ADD223 pKa = 3.12KK224 pKa = 10.76GVRR227 pKa = 11.84VQASLNNVADD237 pKa = 4.36DD238 pKa = 5.19GIAGEE243 pKa = 4.84QEE245 pKa = 3.99NLPNDD250 pKa = 3.36VEE252 pKa = 4.81DD253 pKa = 3.86IYY255 pKa = 11.33GGRR258 pKa = 11.84SADD261 pKa = 3.41VLTGNDD267 pKa = 4.18ADD269 pKa = 3.83NTLNGGPGGGDD280 pKa = 2.97ALYY283 pKa = 11.03GNGGSDD289 pKa = 3.19TLYY292 pKa = 10.92GGSGVDD298 pKa = 4.68KK299 pKa = 11.01LYY301 pKa = 11.4GDD303 pKa = 4.44YY304 pKa = 10.51KK305 pKa = 10.86DD306 pKa = 4.47SAMTYY311 pKa = 10.51GNDD314 pKa = 3.46YY315 pKa = 10.79LYY317 pKa = 11.31GGADD321 pKa = 3.47GDD323 pKa = 4.46LLVGGWGIDD332 pKa = 3.17HH333 pKa = 6.31SRR335 pKa = 11.84EE336 pKa = 3.61YY337 pKa = 10.91DD338 pKa = 3.4YY339 pKa = 10.56DD340 pKa = 3.86TGTDD344 pKa = 3.09TCQSVEE350 pKa = 4.32SEE352 pKa = 4.2DD353 pKa = 3.72HH354 pKa = 6.97DD355 pKa = 3.67KK356 pKa = 11.73CEE358 pKa = 5.72AVLPWPP364 pKa = 4.3
MM1 pKa = 7.17NPIVPTAKK9 pKa = 10.18RR10 pKa = 11.84GIGAAALAAGMVLASPITAGAAAAPPATYY39 pKa = 10.61ASVVGDD45 pKa = 3.68TLNVDD50 pKa = 3.92GNYY53 pKa = 10.01EE54 pKa = 4.41SNHH57 pKa = 4.6ITITPAAYY65 pKa = 9.91GRR67 pKa = 11.84VRR69 pKa = 11.84IADD72 pKa = 3.84ANNPTLPGGDD82 pKa = 4.48CIDD85 pKa = 3.65ITDD88 pKa = 4.32HH89 pKa = 6.57EE90 pKa = 5.07VEE92 pKa = 5.0CGLSGTPTGLDD103 pKa = 3.19VDD105 pKa = 4.05GHH107 pKa = 6.32EE108 pKa = 4.85GADD111 pKa = 3.69TIINLLPTSNKK122 pKa = 7.47TQARR126 pKa = 11.84LHH128 pKa = 6.36GGSGNDD134 pKa = 3.51VIYY137 pKa = 10.44GGPGVQWLNGGLSPDD152 pKa = 4.5DD153 pKa = 3.74LAANQGRR160 pKa = 11.84PDD162 pKa = 3.52TGNDD166 pKa = 3.25KK167 pKa = 11.24LFGGCAQEE175 pKa = 4.76CADD178 pKa = 5.29SGDD181 pKa = 4.04LLEE184 pKa = 5.95GSDD187 pKa = 4.59GNDD190 pKa = 3.36GLTGGPGNDD199 pKa = 4.29DD200 pKa = 3.43LRR202 pKa = 11.84GGIGVDD208 pKa = 3.85TYY210 pKa = 11.7AGGSGRR216 pKa = 11.84DD217 pKa = 3.45VVSYY221 pKa = 10.78ADD223 pKa = 3.12KK224 pKa = 10.76GVRR227 pKa = 11.84VQASLNNVADD237 pKa = 4.36DD238 pKa = 5.19GIAGEE243 pKa = 4.84QEE245 pKa = 3.99NLPNDD250 pKa = 3.36VEE252 pKa = 4.81DD253 pKa = 3.86IYY255 pKa = 11.33GGRR258 pKa = 11.84SADD261 pKa = 3.41VLTGNDD267 pKa = 4.18ADD269 pKa = 3.83NTLNGGPGGGDD280 pKa = 2.97ALYY283 pKa = 11.03GNGGSDD289 pKa = 3.19TLYY292 pKa = 10.92GGSGVDD298 pKa = 4.68KK299 pKa = 11.01LYY301 pKa = 11.4GDD303 pKa = 4.44YY304 pKa = 10.51KK305 pKa = 10.86DD306 pKa = 4.47SAMTYY311 pKa = 10.51GNDD314 pKa = 3.46YY315 pKa = 10.79LYY317 pKa = 11.31GGADD321 pKa = 3.47GDD323 pKa = 4.46LLVGGWGIDD332 pKa = 3.17HH333 pKa = 6.31SRR335 pKa = 11.84EE336 pKa = 3.61YY337 pKa = 10.91DD338 pKa = 3.4YY339 pKa = 10.56DD340 pKa = 3.86TGTDD344 pKa = 3.09TCQSVEE350 pKa = 4.32SEE352 pKa = 4.2DD353 pKa = 3.72HH354 pKa = 6.97DD355 pKa = 3.67KK356 pKa = 11.73CEE358 pKa = 5.72AVLPWPP364 pKa = 4.3
Molecular weight: 36.86 kDa
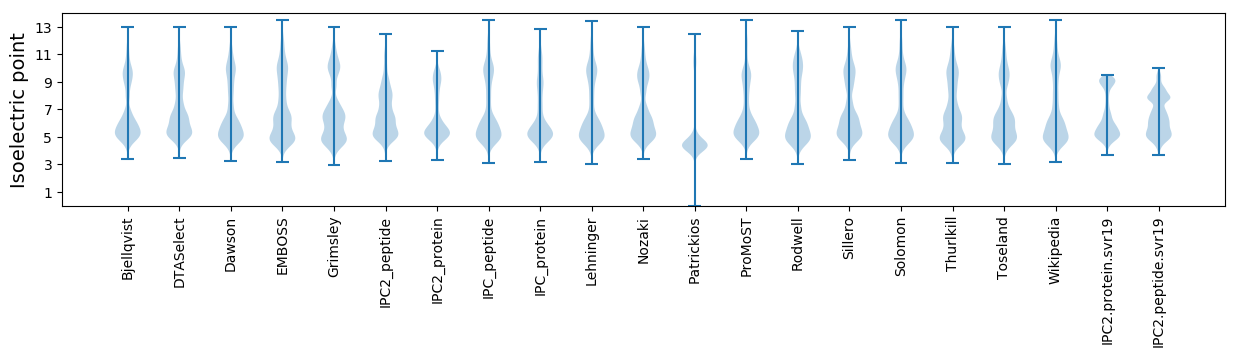
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H6EZP3|A0A1H6EZP3_9ACTN Transport permease protein OS=Nonomuraea solani OX=1144553 GN=SAMN05444920_13610 PE=3 SV=1
MM1 pKa = 8.01RR2 pKa = 11.84GRR4 pKa = 11.84ARR6 pKa = 11.84LRR8 pKa = 11.84ALLRR12 pKa = 11.84VLGRR16 pKa = 11.84MPSPARR22 pKa = 11.84SGPRR26 pKa = 11.84RR27 pKa = 11.84SGCLGPYY34 pKa = 9.14TSGTRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84TPLSLGRR50 pKa = 11.84LRR52 pKa = 11.84SLRR55 pKa = 11.84RR56 pKa = 11.84VTPRR60 pKa = 11.84ARR62 pKa = 11.84PRR64 pKa = 11.84CPQRR68 pKa = 11.84LGRR71 pKa = 11.84PAPRR75 pKa = 11.84LTRR78 pKa = 11.84RR79 pKa = 11.84APAPAAPRR87 pKa = 11.84PNQAPHH93 pKa = 6.9PPNRR97 pKa = 11.84PPPAPLRR104 pKa = 11.84PPSRR108 pKa = 11.84ALHH111 pKa = 5.84LPSRR115 pKa = 11.84APPARR120 pKa = 11.84LPLRR124 pKa = 11.84RR125 pKa = 11.84APLVARR131 pKa = 11.84RR132 pKa = 11.84PPSLVLPPGLRR143 pKa = 11.84PPNRR147 pKa = 11.84ALRR150 pKa = 11.84LPSRR154 pKa = 11.84APVVVPHH161 pKa = 6.88PSRR164 pKa = 11.84APLMAPCPRR173 pKa = 11.84QLVLMAPRR181 pKa = 11.84PRR183 pKa = 11.84RR184 pKa = 11.84APLMVPRR191 pKa = 11.84PRR193 pKa = 11.84RR194 pKa = 11.84PVLRR198 pKa = 11.84GRR200 pKa = 11.84PRR202 pKa = 11.84RR203 pKa = 11.84APALPVGRR211 pKa = 11.84GRR213 pKa = 11.84LRR215 pKa = 11.84RR216 pKa = 11.84PPRR219 pKa = 11.84TSPRR223 pKa = 11.84PSPASAPSRR232 pKa = 11.84PAARR236 pKa = 11.84NRR238 pKa = 11.84PVGLPRR244 pKa = 11.84QVRR247 pKa = 11.84ARR249 pKa = 11.84ARR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84ACDD256 pKa = 3.41RR257 pKa = 11.84SPPAALRR264 pKa = 11.84ARR266 pKa = 11.84PGPNRR271 pKa = 11.84RR272 pKa = 11.84ATLRR276 pKa = 11.84PTPRR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84ARR284 pKa = 11.84PSQAA288 pKa = 3.03
MM1 pKa = 8.01RR2 pKa = 11.84GRR4 pKa = 11.84ARR6 pKa = 11.84LRR8 pKa = 11.84ALLRR12 pKa = 11.84VLGRR16 pKa = 11.84MPSPARR22 pKa = 11.84SGPRR26 pKa = 11.84RR27 pKa = 11.84SGCLGPYY34 pKa = 9.14TSGTRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84TPLSLGRR50 pKa = 11.84LRR52 pKa = 11.84SLRR55 pKa = 11.84RR56 pKa = 11.84VTPRR60 pKa = 11.84ARR62 pKa = 11.84PRR64 pKa = 11.84CPQRR68 pKa = 11.84LGRR71 pKa = 11.84PAPRR75 pKa = 11.84LTRR78 pKa = 11.84RR79 pKa = 11.84APAPAAPRR87 pKa = 11.84PNQAPHH93 pKa = 6.9PPNRR97 pKa = 11.84PPPAPLRR104 pKa = 11.84PPSRR108 pKa = 11.84ALHH111 pKa = 5.84LPSRR115 pKa = 11.84APPARR120 pKa = 11.84LPLRR124 pKa = 11.84RR125 pKa = 11.84APLVARR131 pKa = 11.84RR132 pKa = 11.84PPSLVLPPGLRR143 pKa = 11.84PPNRR147 pKa = 11.84ALRR150 pKa = 11.84LPSRR154 pKa = 11.84APVVVPHH161 pKa = 6.88PSRR164 pKa = 11.84APLMAPCPRR173 pKa = 11.84QLVLMAPRR181 pKa = 11.84PRR183 pKa = 11.84RR184 pKa = 11.84APLMVPRR191 pKa = 11.84PRR193 pKa = 11.84RR194 pKa = 11.84PVLRR198 pKa = 11.84GRR200 pKa = 11.84PRR202 pKa = 11.84RR203 pKa = 11.84APALPVGRR211 pKa = 11.84GRR213 pKa = 11.84LRR215 pKa = 11.84RR216 pKa = 11.84PPRR219 pKa = 11.84TSPRR223 pKa = 11.84PSPASAPSRR232 pKa = 11.84PAARR236 pKa = 11.84NRR238 pKa = 11.84PVGLPRR244 pKa = 11.84QVRR247 pKa = 11.84ARR249 pKa = 11.84ARR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84ACDD256 pKa = 3.41RR257 pKa = 11.84SPPAALRR264 pKa = 11.84ARR266 pKa = 11.84PGPNRR271 pKa = 11.84RR272 pKa = 11.84ATLRR276 pKa = 11.84PTPRR280 pKa = 11.84RR281 pKa = 11.84RR282 pKa = 11.84ARR284 pKa = 11.84PSQAA288 pKa = 3.03
Molecular weight: 31.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3978721 |
27 |
4145 |
328.7 |
35.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.182 ± 0.032 | 0.765 ± 0.006 |
5.781 ± 0.018 | 5.567 ± 0.022 |
2.877 ± 0.014 | 9.371 ± 0.023 |
2.241 ± 0.01 | 3.62 ± 0.013 |
2.1 ± 0.017 | 10.685 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.9 ± 0.009 | 1.874 ± 0.014 |
6.001 ± 0.018 | 2.715 ± 0.012 |
8.083 ± 0.029 | 4.976 ± 0.017 |
6.018 ± 0.023 | 8.426 ± 0.021 |
1.614 ± 0.01 | 2.204 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
