
Actinomadura sp. J1-007
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Thermomonosporaceae; Actinomadura; unclassified Actinomadura
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
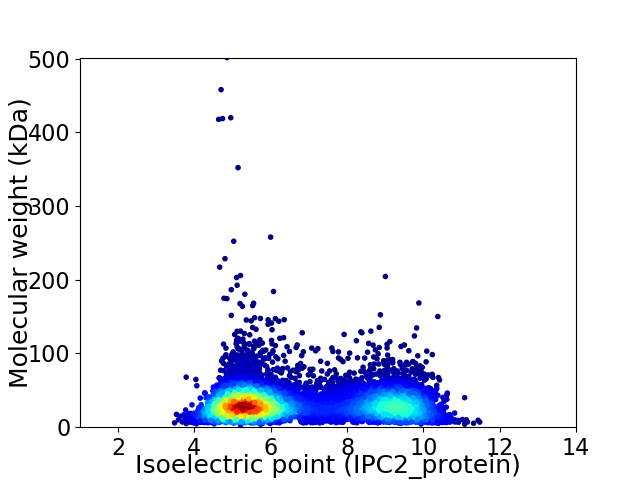
Virtual 2D-PAGE plot for 8101 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I4PQL0|A0A6I4PQL0_9ACTN Helix-turn-helix domain-containing protein OS=Actinomadura sp. J1-007 OX=2661913 GN=GEV43_24405 PE=4 SV=1
MM1 pKa = 6.15TTYY4 pKa = 10.39IVVFGLIVVVVLVVVLVALGMRR26 pKa = 11.84ASRR29 pKa = 11.84AGQDD33 pKa = 3.48DD34 pKa = 5.38DD35 pKa = 5.87DD36 pKa = 5.3WDD38 pKa = 4.68DD39 pKa = 3.73QPQPQPRR46 pKa = 11.84GRR48 pKa = 11.84RR49 pKa = 11.84GVPAEE54 pKa = 5.28DD55 pKa = 3.54MGQDD59 pKa = 3.53AMGGGYY65 pKa = 10.6DD66 pKa = 3.68NGYY69 pKa = 10.94DD70 pKa = 3.5EE71 pKa = 5.88AGYY74 pKa = 10.31DD75 pKa = 3.28AAGYY79 pKa = 10.44DD80 pKa = 3.82GAQYY84 pKa = 11.13GPGSADD90 pKa = 2.84PGYY93 pKa = 10.65DD94 pKa = 2.67RR95 pKa = 11.84RR96 pKa = 11.84VAGAPGGGQGGPGGPQGGGPLAAPAAAPAPPPPAPAGPASDD137 pKa = 4.14EE138 pKa = 4.48MEE140 pKa = 5.67DD141 pKa = 3.45DD142 pKa = 5.39DD143 pKa = 4.41YY144 pKa = 11.55WATITFDD151 pKa = 3.31KK152 pKa = 11.02PKK154 pKa = 10.48FPWQHH159 pKa = 6.61DD160 pKa = 4.04GQPEE164 pKa = 3.87RR165 pKa = 11.84SEE167 pKa = 5.02AEE169 pKa = 4.02MAADD173 pKa = 4.2PLNNPQAPAAEE184 pKa = 4.26QHH186 pKa = 6.36APPAAEE192 pKa = 4.85PPNLTQPVPMGADD205 pKa = 3.38LHH207 pKa = 6.88ALGGVSGPPPAPQGPGGPGGPGGMGTGPQPSGTNPFPGGSPFGGDD252 pKa = 3.4PSATANDD259 pKa = 5.98GIPADD264 pKa = 3.92IGGHH268 pKa = 4.92GQQGPYY274 pKa = 10.68GLGGPGGPDD283 pKa = 2.93QPVYY287 pKa = 10.84GHH289 pKa = 6.09EE290 pKa = 4.02QQPSYY295 pKa = 10.74GAPDD299 pKa = 3.62PSGYY303 pKa = 10.57GGQDD307 pKa = 3.03PSGYY311 pKa = 10.12GQEE314 pKa = 4.09QPSYY318 pKa = 10.72GGDD321 pKa = 3.31PSSPFGVPEE330 pKa = 3.74QQPAYY335 pKa = 10.43GSAQDD340 pKa = 3.45QPSYY344 pKa = 11.27GGAQDD349 pKa = 3.37QPAYY353 pKa = 10.18GGQPEE358 pKa = 4.22QPGPYY363 pKa = 9.91GGGQDD368 pKa = 3.36QPSYY372 pKa = 11.26GDD374 pKa = 3.71YY375 pKa = 11.26GSDD378 pKa = 3.53PLGSRR383 pKa = 11.84PAAAASPPPPPQGRR397 pKa = 11.84PDD399 pKa = 3.78AFDD402 pKa = 4.0MPLSPPAPNAPDD414 pKa = 3.77PRR416 pKa = 11.84ASDD419 pKa = 3.87PLGLPLGRR427 pKa = 11.84TEE429 pKa = 4.89EE430 pKa = 4.3PAPPPAPPAPRR441 pKa = 11.84PAGSDD446 pKa = 3.23TDD448 pKa = 3.61NHH450 pKa = 6.45KK451 pKa = 11.04LPTVDD456 pKa = 5.16EE457 pKa = 4.24LLQRR461 pKa = 11.84IQTDD465 pKa = 3.71RR466 pKa = 11.84QRR468 pKa = 11.84TSGPSDD474 pKa = 2.91QGGSYY479 pKa = 10.87GGGSLNDD486 pKa = 4.02PLGDD490 pKa = 3.79PLGSGSFGTGGGAGIGGSSTGPWAPPPSSGPPSGQGGYY528 pKa = 10.62DD529 pKa = 3.48SDD531 pKa = 4.44PAGSTMSYY539 pKa = 10.85GGGLPAGGSGYY550 pKa = 10.9GQGSQNDD557 pKa = 5.15GYY559 pKa = 9.53PASPAYY565 pKa = 10.12GDD567 pKa = 3.39SSRR570 pKa = 11.84YY571 pKa = 9.75DD572 pKa = 3.48DD573 pKa = 4.47PLGGSGRR580 pKa = 11.84DD581 pKa = 3.39SYY583 pKa = 12.1GSGQGGGMSPAPTGEE598 pKa = 3.86PGRR601 pKa = 11.84YY602 pKa = 9.29GDD604 pKa = 4.93FSGSSYY610 pKa = 11.42NGADD614 pKa = 3.9PLSAPSDD621 pKa = 3.68PGGSGGNYY629 pKa = 9.86DD630 pKa = 3.96PNATQAYY637 pKa = 8.55GSGYY641 pKa = 9.79MNQQSGGYY649 pKa = 7.48QAPSGGDD656 pKa = 3.12TGPYY660 pKa = 8.77GSSYY664 pKa = 10.05PGSDD668 pKa = 3.27QRR670 pKa = 11.84QPDD673 pKa = 3.36DD674 pKa = 3.64WEE676 pKa = 3.72NHH678 pKa = 4.52RR679 pKa = 11.84DD680 pKa = 3.59YY681 pKa = 11.29RR682 pKa = 11.84RR683 pKa = 3.16
MM1 pKa = 6.15TTYY4 pKa = 10.39IVVFGLIVVVVLVVVLVALGMRR26 pKa = 11.84ASRR29 pKa = 11.84AGQDD33 pKa = 3.48DD34 pKa = 5.38DD35 pKa = 5.87DD36 pKa = 5.3WDD38 pKa = 4.68DD39 pKa = 3.73QPQPQPRR46 pKa = 11.84GRR48 pKa = 11.84RR49 pKa = 11.84GVPAEE54 pKa = 5.28DD55 pKa = 3.54MGQDD59 pKa = 3.53AMGGGYY65 pKa = 10.6DD66 pKa = 3.68NGYY69 pKa = 10.94DD70 pKa = 3.5EE71 pKa = 5.88AGYY74 pKa = 10.31DD75 pKa = 3.28AAGYY79 pKa = 10.44DD80 pKa = 3.82GAQYY84 pKa = 11.13GPGSADD90 pKa = 2.84PGYY93 pKa = 10.65DD94 pKa = 2.67RR95 pKa = 11.84RR96 pKa = 11.84VAGAPGGGQGGPGGPQGGGPLAAPAAAPAPPPPAPAGPASDD137 pKa = 4.14EE138 pKa = 4.48MEE140 pKa = 5.67DD141 pKa = 3.45DD142 pKa = 5.39DD143 pKa = 4.41YY144 pKa = 11.55WATITFDD151 pKa = 3.31KK152 pKa = 11.02PKK154 pKa = 10.48FPWQHH159 pKa = 6.61DD160 pKa = 4.04GQPEE164 pKa = 3.87RR165 pKa = 11.84SEE167 pKa = 5.02AEE169 pKa = 4.02MAADD173 pKa = 4.2PLNNPQAPAAEE184 pKa = 4.26QHH186 pKa = 6.36APPAAEE192 pKa = 4.85PPNLTQPVPMGADD205 pKa = 3.38LHH207 pKa = 6.88ALGGVSGPPPAPQGPGGPGGPGGMGTGPQPSGTNPFPGGSPFGGDD252 pKa = 3.4PSATANDD259 pKa = 5.98GIPADD264 pKa = 3.92IGGHH268 pKa = 4.92GQQGPYY274 pKa = 10.68GLGGPGGPDD283 pKa = 2.93QPVYY287 pKa = 10.84GHH289 pKa = 6.09EE290 pKa = 4.02QQPSYY295 pKa = 10.74GAPDD299 pKa = 3.62PSGYY303 pKa = 10.57GGQDD307 pKa = 3.03PSGYY311 pKa = 10.12GQEE314 pKa = 4.09QPSYY318 pKa = 10.72GGDD321 pKa = 3.31PSSPFGVPEE330 pKa = 3.74QQPAYY335 pKa = 10.43GSAQDD340 pKa = 3.45QPSYY344 pKa = 11.27GGAQDD349 pKa = 3.37QPAYY353 pKa = 10.18GGQPEE358 pKa = 4.22QPGPYY363 pKa = 9.91GGGQDD368 pKa = 3.36QPSYY372 pKa = 11.26GDD374 pKa = 3.71YY375 pKa = 11.26GSDD378 pKa = 3.53PLGSRR383 pKa = 11.84PAAAASPPPPPQGRR397 pKa = 11.84PDD399 pKa = 3.78AFDD402 pKa = 4.0MPLSPPAPNAPDD414 pKa = 3.77PRR416 pKa = 11.84ASDD419 pKa = 3.87PLGLPLGRR427 pKa = 11.84TEE429 pKa = 4.89EE430 pKa = 4.3PAPPPAPPAPRR441 pKa = 11.84PAGSDD446 pKa = 3.23TDD448 pKa = 3.61NHH450 pKa = 6.45KK451 pKa = 11.04LPTVDD456 pKa = 5.16EE457 pKa = 4.24LLQRR461 pKa = 11.84IQTDD465 pKa = 3.71RR466 pKa = 11.84QRR468 pKa = 11.84TSGPSDD474 pKa = 2.91QGGSYY479 pKa = 10.87GGGSLNDD486 pKa = 4.02PLGDD490 pKa = 3.79PLGSGSFGTGGGAGIGGSSTGPWAPPPSSGPPSGQGGYY528 pKa = 10.62DD529 pKa = 3.48SDD531 pKa = 4.44PAGSTMSYY539 pKa = 10.85GGGLPAGGSGYY550 pKa = 10.9GQGSQNDD557 pKa = 5.15GYY559 pKa = 9.53PASPAYY565 pKa = 10.12GDD567 pKa = 3.39SSRR570 pKa = 11.84YY571 pKa = 9.75DD572 pKa = 3.48DD573 pKa = 4.47PLGGSGRR580 pKa = 11.84DD581 pKa = 3.39SYY583 pKa = 12.1GSGQGGGMSPAPTGEE598 pKa = 3.86PGRR601 pKa = 11.84YY602 pKa = 9.29GDD604 pKa = 4.93FSGSSYY610 pKa = 11.42NGADD614 pKa = 3.9PLSAPSDD621 pKa = 3.68PGGSGGNYY629 pKa = 9.86DD630 pKa = 3.96PNATQAYY637 pKa = 8.55GSGYY641 pKa = 9.79MNQQSGGYY649 pKa = 7.48QAPSGGDD656 pKa = 3.12TGPYY660 pKa = 8.77GSSYY664 pKa = 10.05PGSDD668 pKa = 3.27QRR670 pKa = 11.84QPDD673 pKa = 3.36DD674 pKa = 3.64WEE676 pKa = 3.72NHH678 pKa = 4.52RR679 pKa = 11.84DD680 pKa = 3.59YY681 pKa = 11.29RR682 pKa = 11.84RR683 pKa = 3.16
Molecular weight: 67.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I4PMR3|A0A6I4PMR3_9ACTN D-alanine--D-alanine ligase OS=Actinomadura sp. J1-007 OX=2661913 GN=ddl PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.34KK15 pKa = 10.47KK16 pKa = 8.84HH17 pKa = 5.5GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSGRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.86GRR40 pKa = 11.84ARR42 pKa = 11.84IAVV45 pKa = 3.5
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.34KK15 pKa = 10.47KK16 pKa = 8.84HH17 pKa = 5.5GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSGRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.86GRR40 pKa = 11.84ARR42 pKa = 11.84IAVV45 pKa = 3.5
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2547833 |
29 |
4770 |
314.5 |
33.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.301 ± 0.045 | 0.793 ± 0.007 |
5.912 ± 0.019 | 5.47 ± 0.025 |
2.745 ± 0.014 | 9.99 ± 0.027 |
2.135 ± 0.012 | 3.078 ± 0.016 |
1.873 ± 0.022 | 10.175 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.783 ± 0.01 | 1.673 ± 0.013 |
6.465 ± 0.026 | 2.307 ± 0.014 |
9.014 ± 0.034 | 4.818 ± 0.021 |
5.5 ± 0.023 | 8.469 ± 0.025 |
1.512 ± 0.011 | 1.987 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
