
Magnetospira sp. (strain QH-2) (Marine magnetic spirillum (strain QH-2))
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Thalassospiraceae; Magnetospira; unclassified Magnetospira
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
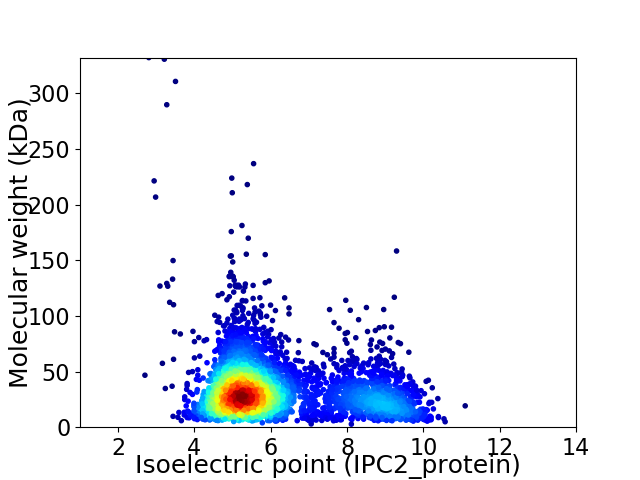
Virtual 2D-PAGE plot for 3700 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
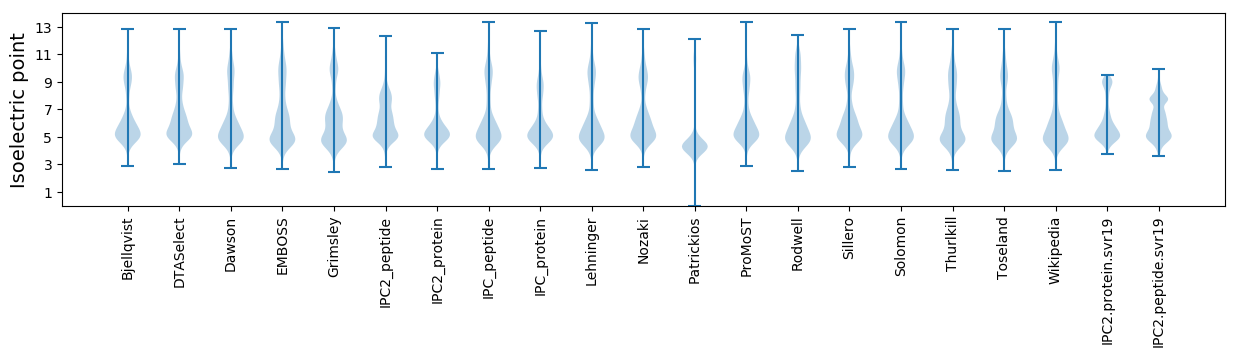
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W6KPV5|W6KPV5_MAGSQ 2-oxo-carboxylic acid reductase (Glyoxalate reductase) (2-ketoaldonate reductase) OS=Magnetospira sp. (strain QH-2) OX=1288970 GN=MGMAQ_3928 PE=3 SV=1
MM1 pKa = 7.48KK2 pKa = 10.29KK3 pKa = 10.36VLLTTTALVVAGAFASQAQAADD25 pKa = 4.41PIKK28 pKa = 10.74LSVGGHH34 pKa = 4.37MNQWIMFADD43 pKa = 3.78SDD45 pKa = 4.67KK46 pKa = 10.65ATQTGGVDD54 pKa = 3.39TLSDD58 pKa = 3.56TEE60 pKa = 4.9VYY62 pKa = 10.61FKK64 pKa = 11.45GDD66 pKa = 3.27TTLEE70 pKa = 4.18NGLKK74 pKa = 10.0VSVVIQLEE82 pKa = 4.39GEE84 pKa = 3.98EE85 pKa = 4.12DD86 pKa = 3.64TATNNADD93 pKa = 3.32EE94 pKa = 4.46QYY96 pKa = 11.53VVISSDD102 pKa = 3.18SLGTLRR108 pKa = 11.84VGDD111 pKa = 4.59KK112 pKa = 8.58EE113 pKa = 4.3TALATIKK120 pKa = 10.29IGSPAVGYY128 pKa = 9.93GGPTDD133 pKa = 4.4ADD135 pKa = 3.63DD136 pKa = 4.1VVAPAFAGASDD147 pKa = 3.48QFANAEE153 pKa = 4.14FVDD156 pKa = 4.98DD157 pKa = 5.56DD158 pKa = 4.9DD159 pKa = 4.96NQVTYY164 pKa = 10.51LSPSFAGFQVGGSWVPHH181 pKa = 6.07FAAGDD186 pKa = 3.79AQGGPNTNPVGGNNDD201 pKa = 3.66GGWGVGAIYY210 pKa = 10.55SADD213 pKa = 3.44YY214 pKa = 11.01DD215 pKa = 4.32GIGVGVSGGYY225 pKa = 10.13AYY227 pKa = 10.49HH228 pKa = 7.85DD229 pKa = 4.31AGADD233 pKa = 2.92IWQAGAKK240 pKa = 9.99VSFAGFTVSGGWQRR254 pKa = 11.84TDD256 pKa = 2.85VHH258 pKa = 6.48EE259 pKa = 4.39VQLGNTNNALSVDD272 pKa = 3.89GVEE275 pKa = 4.41GTGWEE280 pKa = 4.2AGVSYY285 pKa = 9.24ATGPYY290 pKa = 9.89AVSLVYY296 pKa = 10.32GAGEE300 pKa = 5.01DD301 pKa = 3.67EE302 pKa = 4.59DD303 pKa = 5.18TYY305 pKa = 11.98ADD307 pKa = 3.63TDD309 pKa = 3.77KK310 pKa = 11.83DD311 pKa = 3.93EE312 pKa = 4.86LKK314 pKa = 10.67VWAVGANYY322 pKa = 11.0SMGAGVMMKK331 pKa = 9.35GTIYY335 pKa = 10.38HH336 pKa = 6.99ADD338 pKa = 3.55YY339 pKa = 11.17NGEE342 pKa = 4.21TNTTGVGGDD351 pKa = 3.72TASGWGGIVGMAISFF366 pKa = 3.8
MM1 pKa = 7.48KK2 pKa = 10.29KK3 pKa = 10.36VLLTTTALVVAGAFASQAQAADD25 pKa = 4.41PIKK28 pKa = 10.74LSVGGHH34 pKa = 4.37MNQWIMFADD43 pKa = 3.78SDD45 pKa = 4.67KK46 pKa = 10.65ATQTGGVDD54 pKa = 3.39TLSDD58 pKa = 3.56TEE60 pKa = 4.9VYY62 pKa = 10.61FKK64 pKa = 11.45GDD66 pKa = 3.27TTLEE70 pKa = 4.18NGLKK74 pKa = 10.0VSVVIQLEE82 pKa = 4.39GEE84 pKa = 3.98EE85 pKa = 4.12DD86 pKa = 3.64TATNNADD93 pKa = 3.32EE94 pKa = 4.46QYY96 pKa = 11.53VVISSDD102 pKa = 3.18SLGTLRR108 pKa = 11.84VGDD111 pKa = 4.59KK112 pKa = 8.58EE113 pKa = 4.3TALATIKK120 pKa = 10.29IGSPAVGYY128 pKa = 9.93GGPTDD133 pKa = 4.4ADD135 pKa = 3.63DD136 pKa = 4.1VVAPAFAGASDD147 pKa = 3.48QFANAEE153 pKa = 4.14FVDD156 pKa = 4.98DD157 pKa = 5.56DD158 pKa = 4.9DD159 pKa = 4.96NQVTYY164 pKa = 10.51LSPSFAGFQVGGSWVPHH181 pKa = 6.07FAAGDD186 pKa = 3.79AQGGPNTNPVGGNNDD201 pKa = 3.66GGWGVGAIYY210 pKa = 10.55SADD213 pKa = 3.44YY214 pKa = 11.01DD215 pKa = 4.32GIGVGVSGGYY225 pKa = 10.13AYY227 pKa = 10.49HH228 pKa = 7.85DD229 pKa = 4.31AGADD233 pKa = 2.92IWQAGAKK240 pKa = 9.99VSFAGFTVSGGWQRR254 pKa = 11.84TDD256 pKa = 2.85VHH258 pKa = 6.48EE259 pKa = 4.39VQLGNTNNALSVDD272 pKa = 3.89GVEE275 pKa = 4.41GTGWEE280 pKa = 4.2AGVSYY285 pKa = 9.24ATGPYY290 pKa = 9.89AVSLVYY296 pKa = 10.32GAGEE300 pKa = 5.01DD301 pKa = 3.67EE302 pKa = 4.59DD303 pKa = 5.18TYY305 pKa = 11.98ADD307 pKa = 3.63TDD309 pKa = 3.77KK310 pKa = 11.83DD311 pKa = 3.93EE312 pKa = 4.86LKK314 pKa = 10.67VWAVGANYY322 pKa = 11.0SMGAGVMMKK331 pKa = 9.35GTIYY335 pKa = 10.38HH336 pKa = 6.99ADD338 pKa = 3.55YY339 pKa = 11.17NGEE342 pKa = 4.21TNTTGVGGDD351 pKa = 3.72TASGWGGIVGMAISFF366 pKa = 3.8
Molecular weight: 37.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W6K585|W6K585_MAGSQ Uncharacterized protein OS=Magnetospira sp. (strain QH-2) OX=1288970 GN=MGMAQ_0348 PE=4 SV=1
MM1 pKa = 8.11RR2 pKa = 11.84IPGTRR7 pKa = 11.84LSLLLVALPSALLAPLLGLALLALRR32 pKa = 11.84RR33 pKa = 11.84GSALPISLLALRR45 pKa = 11.84RR46 pKa = 11.84WATLPTALLATLLGTALLPTGGRR69 pKa = 11.84TTLPATLLAASPRR82 pKa = 11.84AALPLSLRR90 pKa = 11.84ALGRR94 pKa = 11.84RR95 pKa = 11.84PTLPVALLALRR106 pKa = 11.84WRR108 pKa = 11.84TPLPVSRR115 pKa = 11.84LTALPGTALPLSLLSSRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84PALPVALLALRR145 pKa = 11.84WRR147 pKa = 11.84PPLPLPWRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84VCLLLRR163 pKa = 11.84HH164 pKa = 5.42WTRR167 pKa = 11.84PPKK170 pKa = 10.66NGGKK174 pKa = 10.05RR175 pKa = 11.84GGGEE179 pKa = 3.66
MM1 pKa = 8.11RR2 pKa = 11.84IPGTRR7 pKa = 11.84LSLLLVALPSALLAPLLGLALLALRR32 pKa = 11.84RR33 pKa = 11.84GSALPISLLALRR45 pKa = 11.84RR46 pKa = 11.84WATLPTALLATLLGTALLPTGGRR69 pKa = 11.84TTLPATLLAASPRR82 pKa = 11.84AALPLSLRR90 pKa = 11.84ALGRR94 pKa = 11.84RR95 pKa = 11.84PTLPVALLALRR106 pKa = 11.84WRR108 pKa = 11.84TPLPVSRR115 pKa = 11.84LTALPGTALPLSLLSSRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84PALPVALLALRR145 pKa = 11.84WRR147 pKa = 11.84PPLPLPWRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84VCLLLRR163 pKa = 11.84HH164 pKa = 5.42WTRR167 pKa = 11.84PPKK170 pKa = 10.66NGGKK174 pKa = 10.05RR175 pKa = 11.84GGGEE179 pKa = 3.66
Molecular weight: 19.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1196687 |
23 |
3296 |
323.4 |
35.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.641 ± 0.052 | 0.914 ± 0.016 |
6.445 ± 0.055 | 6.185 ± 0.039 |
3.629 ± 0.025 | 8.54 ± 0.052 |
2.274 ± 0.021 | 5.038 ± 0.035 |
3.893 ± 0.04 | 10.329 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.751 ± 0.022 | 2.843 ± 0.023 |
5.073 ± 0.036 | 3.327 ± 0.025 |
6.568 ± 0.043 | 5.281 ± 0.029 |
5.367 ± 0.037 | 7.221 ± 0.033 |
1.357 ± 0.015 | 2.326 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
