
Roseibium sp. TrichSKD4
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Stappiaceae; Roseibium; unclassified Roseibium
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
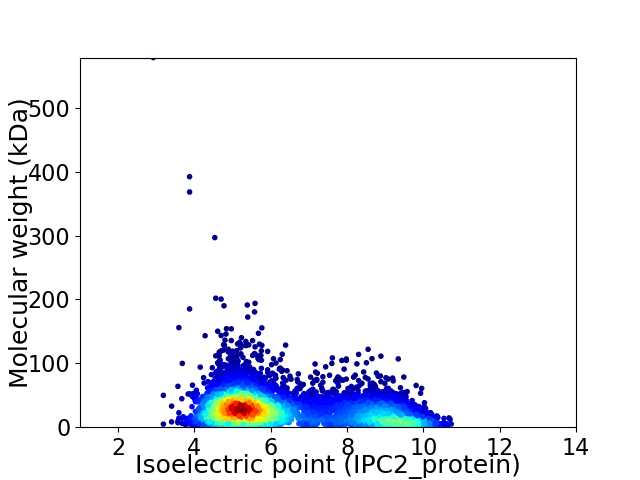
Virtual 2D-PAGE plot for 5884 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2CIE0|E2CIE0_9RHOB Ribonuclease T2 OS=Roseibium sp. TrichSKD4 OX=744980 GN=TRICHSKD4_2842 PE=3 SV=1
MM1 pKa = 7.69IGTNDD6 pKa = 3.97DD7 pKa = 4.95DD8 pKa = 3.99ILNGTVHH15 pKa = 6.91SDD17 pKa = 3.65VISGRR22 pKa = 11.84FGDD25 pKa = 4.13DD26 pKa = 3.01TLIGANGNDD35 pKa = 3.98EE36 pKa = 4.31VWGGTGDD43 pKa = 3.56DD44 pKa = 3.5TLYY47 pKa = 11.3GNNGNDD53 pKa = 3.01ILYY56 pKa = 10.83GSGGPNLVQITSVEE70 pKa = 3.91IGDD73 pKa = 4.75DD74 pKa = 3.91YY75 pKa = 11.19PVSVVFEE82 pKa = 4.55GEE84 pKa = 3.88TAGYY88 pKa = 10.2RR89 pKa = 11.84NTFGYY94 pKa = 10.72YY95 pKa = 9.52KK96 pKa = 10.34VSEE99 pKa = 5.1DD100 pKa = 3.02GTIYY104 pKa = 10.13HH105 pKa = 6.41VEE107 pKa = 4.41VIWPNASLQGSGGNLIQGEE126 pKa = 4.47SRR128 pKa = 11.84EE129 pKa = 4.05FLDD132 pKa = 4.29VQSGDD137 pKa = 3.27QLAFFIISNGYY148 pKa = 8.53SRR150 pKa = 11.84NGGYY154 pKa = 10.66AGIDD158 pKa = 3.65LEE160 pKa = 4.53NGTLEE165 pKa = 4.19FQEE168 pKa = 4.63SDD170 pKa = 3.1GTTATINSTSPRR182 pKa = 11.84LYY184 pKa = 10.81YY185 pKa = 10.02IAADD189 pKa = 3.87GTRR192 pKa = 11.84TMIQTDD198 pKa = 4.41PYY200 pKa = 8.86HH201 pKa = 5.58TAAYY205 pKa = 9.78GDD207 pKa = 4.09TVGLNPDD214 pKa = 3.91GFLHH218 pKa = 5.23TTGVLKK224 pKa = 10.13TDD226 pKa = 3.6AGTITLGFEE235 pKa = 4.06DD236 pKa = 5.82LYY238 pKa = 11.66NGGDD242 pKa = 3.5RR243 pKa = 11.84DD244 pKa = 3.72FDD246 pKa = 4.41DD247 pKa = 4.88SVFTVDD253 pKa = 4.39IGVANALVLNAHH265 pKa = 6.08YY266 pKa = 10.42RR267 pKa = 11.84GQNEE271 pKa = 4.6GEE273 pKa = 4.56DD274 pKa = 4.22PDD276 pKa = 4.42TGDD279 pKa = 3.43GEE281 pKa = 4.44NPVVIEE287 pKa = 4.79RR288 pKa = 11.84SDD290 pKa = 3.45NDD292 pKa = 3.2ILYY295 pKa = 10.59GGTGQDD301 pKa = 3.51EE302 pKa = 4.33LHH304 pKa = 6.47GRR306 pKa = 11.84SGDD309 pKa = 3.5DD310 pKa = 2.93TLYY313 pKa = 11.35GNNGEE318 pKa = 4.74DD319 pKa = 4.15DD320 pKa = 3.81LHH322 pKa = 7.4GGSGDD327 pKa = 3.53DD328 pKa = 3.5ALYY331 pKa = 10.85GGSASDD337 pKa = 3.54NLYY340 pKa = 11.13GNSGNDD346 pKa = 3.45TLFGGQGNDD355 pKa = 3.42SLNGNNGDD363 pKa = 4.75DD364 pKa = 3.71ILNGEE369 pKa = 4.47SGNDD373 pKa = 3.4EE374 pKa = 5.03LIGGTGNDD382 pKa = 4.01TLDD385 pKa = 3.91GGHH388 pKa = 6.79GNDD391 pKa = 4.48ILDD394 pKa = 3.87GEE396 pKa = 4.59YY397 pKa = 11.08GNDD400 pKa = 3.62VIKK403 pKa = 10.97GGAGDD408 pKa = 3.61DD409 pKa = 3.84QLRR412 pKa = 11.84GGYY415 pKa = 10.46NNDD418 pKa = 3.37EE419 pKa = 4.36LFGGHH424 pKa = 6.76GNDD427 pKa = 3.66TFYY430 pKa = 11.75GDD432 pKa = 3.47AGNDD436 pKa = 3.36VMHH439 pKa = 7.32GGRR442 pKa = 11.84GVDD445 pKa = 3.32TVDD448 pKa = 3.11YY449 pKa = 10.58SAYY452 pKa = 10.54DD453 pKa = 3.19VDD455 pKa = 4.62LTVSLHH461 pKa = 5.28NKK463 pKa = 8.12KK464 pKa = 10.3AHH466 pKa = 5.42GLEE469 pKa = 4.28IGSDD473 pKa = 3.44TLKK476 pKa = 11.06FIDD479 pKa = 5.28NITSGDD485 pKa = 3.44GDD487 pKa = 4.44DD488 pKa = 4.13FLKK491 pKa = 11.0GSSGDD496 pKa = 3.57NVINGGAGDD505 pKa = 3.74DD506 pKa = 4.27TIRR509 pKa = 11.84GLTGEE514 pKa = 4.49DD515 pKa = 4.06VLTGGAGADD524 pKa = 3.1TFVFRR529 pKa = 11.84TFDD532 pKa = 3.95LNPADD537 pKa = 4.52VITDD541 pKa = 4.3FDD543 pKa = 4.87LLNDD547 pKa = 4.09TLDD550 pKa = 3.84LSHH553 pKa = 7.21LAGGEE558 pKa = 4.3GEE560 pKa = 4.75GFLSSLQALEE570 pKa = 4.35QGDD573 pKa = 4.33GTLLSVDD580 pKa = 3.94LNGDD584 pKa = 3.34GSYY587 pKa = 11.2QDD589 pKa = 3.06ICSLEE594 pKa = 4.62GIGSVSLTTLNNNDD608 pKa = 3.61CFLFF612 pKa = 4.6
MM1 pKa = 7.69IGTNDD6 pKa = 3.97DD7 pKa = 4.95DD8 pKa = 3.99ILNGTVHH15 pKa = 6.91SDD17 pKa = 3.65VISGRR22 pKa = 11.84FGDD25 pKa = 4.13DD26 pKa = 3.01TLIGANGNDD35 pKa = 3.98EE36 pKa = 4.31VWGGTGDD43 pKa = 3.56DD44 pKa = 3.5TLYY47 pKa = 11.3GNNGNDD53 pKa = 3.01ILYY56 pKa = 10.83GSGGPNLVQITSVEE70 pKa = 3.91IGDD73 pKa = 4.75DD74 pKa = 3.91YY75 pKa = 11.19PVSVVFEE82 pKa = 4.55GEE84 pKa = 3.88TAGYY88 pKa = 10.2RR89 pKa = 11.84NTFGYY94 pKa = 10.72YY95 pKa = 9.52KK96 pKa = 10.34VSEE99 pKa = 5.1DD100 pKa = 3.02GTIYY104 pKa = 10.13HH105 pKa = 6.41VEE107 pKa = 4.41VIWPNASLQGSGGNLIQGEE126 pKa = 4.47SRR128 pKa = 11.84EE129 pKa = 4.05FLDD132 pKa = 4.29VQSGDD137 pKa = 3.27QLAFFIISNGYY148 pKa = 8.53SRR150 pKa = 11.84NGGYY154 pKa = 10.66AGIDD158 pKa = 3.65LEE160 pKa = 4.53NGTLEE165 pKa = 4.19FQEE168 pKa = 4.63SDD170 pKa = 3.1GTTATINSTSPRR182 pKa = 11.84LYY184 pKa = 10.81YY185 pKa = 10.02IAADD189 pKa = 3.87GTRR192 pKa = 11.84TMIQTDD198 pKa = 4.41PYY200 pKa = 8.86HH201 pKa = 5.58TAAYY205 pKa = 9.78GDD207 pKa = 4.09TVGLNPDD214 pKa = 3.91GFLHH218 pKa = 5.23TTGVLKK224 pKa = 10.13TDD226 pKa = 3.6AGTITLGFEE235 pKa = 4.06DD236 pKa = 5.82LYY238 pKa = 11.66NGGDD242 pKa = 3.5RR243 pKa = 11.84DD244 pKa = 3.72FDD246 pKa = 4.41DD247 pKa = 4.88SVFTVDD253 pKa = 4.39IGVANALVLNAHH265 pKa = 6.08YY266 pKa = 10.42RR267 pKa = 11.84GQNEE271 pKa = 4.6GEE273 pKa = 4.56DD274 pKa = 4.22PDD276 pKa = 4.42TGDD279 pKa = 3.43GEE281 pKa = 4.44NPVVIEE287 pKa = 4.79RR288 pKa = 11.84SDD290 pKa = 3.45NDD292 pKa = 3.2ILYY295 pKa = 10.59GGTGQDD301 pKa = 3.51EE302 pKa = 4.33LHH304 pKa = 6.47GRR306 pKa = 11.84SGDD309 pKa = 3.5DD310 pKa = 2.93TLYY313 pKa = 11.35GNNGEE318 pKa = 4.74DD319 pKa = 4.15DD320 pKa = 3.81LHH322 pKa = 7.4GGSGDD327 pKa = 3.53DD328 pKa = 3.5ALYY331 pKa = 10.85GGSASDD337 pKa = 3.54NLYY340 pKa = 11.13GNSGNDD346 pKa = 3.45TLFGGQGNDD355 pKa = 3.42SLNGNNGDD363 pKa = 4.75DD364 pKa = 3.71ILNGEE369 pKa = 4.47SGNDD373 pKa = 3.4EE374 pKa = 5.03LIGGTGNDD382 pKa = 4.01TLDD385 pKa = 3.91GGHH388 pKa = 6.79GNDD391 pKa = 4.48ILDD394 pKa = 3.87GEE396 pKa = 4.59YY397 pKa = 11.08GNDD400 pKa = 3.62VIKK403 pKa = 10.97GGAGDD408 pKa = 3.61DD409 pKa = 3.84QLRR412 pKa = 11.84GGYY415 pKa = 10.46NNDD418 pKa = 3.37EE419 pKa = 4.36LFGGHH424 pKa = 6.76GNDD427 pKa = 3.66TFYY430 pKa = 11.75GDD432 pKa = 3.47AGNDD436 pKa = 3.36VMHH439 pKa = 7.32GGRR442 pKa = 11.84GVDD445 pKa = 3.32TVDD448 pKa = 3.11YY449 pKa = 10.58SAYY452 pKa = 10.54DD453 pKa = 3.19VDD455 pKa = 4.62LTVSLHH461 pKa = 5.28NKK463 pKa = 8.12KK464 pKa = 10.3AHH466 pKa = 5.42GLEE469 pKa = 4.28IGSDD473 pKa = 3.44TLKK476 pKa = 11.06FIDD479 pKa = 5.28NITSGDD485 pKa = 3.44GDD487 pKa = 4.44DD488 pKa = 4.13FLKK491 pKa = 11.0GSSGDD496 pKa = 3.57NVINGGAGDD505 pKa = 3.74DD506 pKa = 4.27TIRR509 pKa = 11.84GLTGEE514 pKa = 4.49DD515 pKa = 4.06VLTGGAGADD524 pKa = 3.1TFVFRR529 pKa = 11.84TFDD532 pKa = 3.95LNPADD537 pKa = 4.52VITDD541 pKa = 4.3FDD543 pKa = 4.87LLNDD547 pKa = 4.09TLDD550 pKa = 3.84LSHH553 pKa = 7.21LAGGEE558 pKa = 4.3GEE560 pKa = 4.75GFLSSLQALEE570 pKa = 4.35QGDD573 pKa = 4.33GTLLSVDD580 pKa = 3.94LNGDD584 pKa = 3.34GSYY587 pKa = 11.2QDD589 pKa = 3.06ICSLEE594 pKa = 4.62GIGSVSLTTLNNNDD608 pKa = 3.61CFLFF612 pKa = 4.6
Molecular weight: 63.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2CK55|E2CK55_9RHOB Uncharacterized protein OS=Roseibium sp. TrichSKD4 OX=744980 GN=TRICHSKD4_3471 PE=4 SV=1
MM1 pKa = 7.33QFIADD6 pKa = 4.05WKK8 pKa = 10.21RR9 pKa = 11.84VFAVSLSFWTLVLGFLILLVPEE31 pKa = 3.86AVFRR35 pKa = 11.84FTGADD40 pKa = 3.37TNPYY44 pKa = 9.13LVGWFAVFLFVAGMVGRR61 pKa = 11.84FVKK64 pKa = 10.39QDD66 pKa = 2.86GSKK69 pKa = 9.79RR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 3.67WLRR75 pKa = 11.84ILGVVLGALVIAMALSGQGIAGQMAEE101 pKa = 4.13SRR103 pKa = 11.84EE104 pKa = 4.04AATLKK109 pKa = 10.45IAVPFIAAKK118 pKa = 9.35EE119 pKa = 4.28GKK121 pKa = 9.85RR122 pKa = 11.84NRR124 pKa = 11.84AYY126 pKa = 10.84LDD128 pKa = 3.46VVGVPTICYY137 pKa = 9.66GSTRR141 pKa = 11.84GVKK144 pKa = 10.32LGMVKK149 pKa = 9.72TNAEE153 pKa = 4.09CTALLRR159 pKa = 11.84DD160 pKa = 3.57EE161 pKa = 4.38VAEE164 pKa = 4.08YY165 pKa = 10.79RR166 pKa = 11.84HH167 pKa = 5.98GLHH170 pKa = 7.17PYY172 pKa = 7.71FTKK175 pKa = 8.39TTKK178 pKa = 9.98SRR180 pKa = 11.84RR181 pKa = 11.84LPPSRR186 pKa = 11.84DD187 pKa = 3.12AAFTSLAFNCGIRR200 pKa = 11.84AIGRR204 pKa = 11.84STATRR209 pKa = 11.84RR210 pKa = 11.84LNSGDD215 pKa = 3.03IRR217 pKa = 11.84GACHH221 pKa = 7.64AITWWNKK228 pKa = 8.51AGGRR232 pKa = 11.84VWRR235 pKa = 11.84GLVVRR240 pKa = 11.84RR241 pKa = 11.84SAEE244 pKa = 3.72RR245 pKa = 11.84DD246 pKa = 3.15LCLGNGTT253 pKa = 4.32
MM1 pKa = 7.33QFIADD6 pKa = 4.05WKK8 pKa = 10.21RR9 pKa = 11.84VFAVSLSFWTLVLGFLILLVPEE31 pKa = 3.86AVFRR35 pKa = 11.84FTGADD40 pKa = 3.37TNPYY44 pKa = 9.13LVGWFAVFLFVAGMVGRR61 pKa = 11.84FVKK64 pKa = 10.39QDD66 pKa = 2.86GSKK69 pKa = 9.79RR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 3.67WLRR75 pKa = 11.84ILGVVLGALVIAMALSGQGIAGQMAEE101 pKa = 4.13SRR103 pKa = 11.84EE104 pKa = 4.04AATLKK109 pKa = 10.45IAVPFIAAKK118 pKa = 9.35EE119 pKa = 4.28GKK121 pKa = 9.85RR122 pKa = 11.84NRR124 pKa = 11.84AYY126 pKa = 10.84LDD128 pKa = 3.46VVGVPTICYY137 pKa = 9.66GSTRR141 pKa = 11.84GVKK144 pKa = 10.32LGMVKK149 pKa = 9.72TNAEE153 pKa = 4.09CTALLRR159 pKa = 11.84DD160 pKa = 3.57EE161 pKa = 4.38VAEE164 pKa = 4.08YY165 pKa = 10.79RR166 pKa = 11.84HH167 pKa = 5.98GLHH170 pKa = 7.17PYY172 pKa = 7.71FTKK175 pKa = 8.39TTKK178 pKa = 9.98SRR180 pKa = 11.84RR181 pKa = 11.84LPPSRR186 pKa = 11.84DD187 pKa = 3.12AAFTSLAFNCGIRR200 pKa = 11.84AIGRR204 pKa = 11.84STATRR209 pKa = 11.84RR210 pKa = 11.84LNSGDD215 pKa = 3.03IRR217 pKa = 11.84GACHH221 pKa = 7.64AITWWNKK228 pKa = 8.51AGGRR232 pKa = 11.84VWRR235 pKa = 11.84GLVVRR240 pKa = 11.84RR241 pKa = 11.84SAEE244 pKa = 3.72RR245 pKa = 11.84DD246 pKa = 3.15LCLGNGTT253 pKa = 4.32
Molecular weight: 27.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1621200 |
17 |
6021 |
275.5 |
30.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.578 ± 0.043 | 0.926 ± 0.013 |
5.982 ± 0.05 | 6.313 ± 0.036 |
3.978 ± 0.023 | 8.114 ± 0.057 |
2.046 ± 0.016 | 5.422 ± 0.023 |
4.382 ± 0.033 | 9.9 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.491 ± 0.019 | 3.232 ± 0.025 |
4.702 ± 0.031 | 3.449 ± 0.019 |
6.126 ± 0.033 | 6.115 ± 0.034 |
5.49 ± 0.035 | 7.095 ± 0.027 |
1.316 ± 0.015 | 2.328 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
