
Halobacterium hubeiense
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Halobacterium
Average proteome isoelectric point is 4.88
Get precalculated fractions of proteins
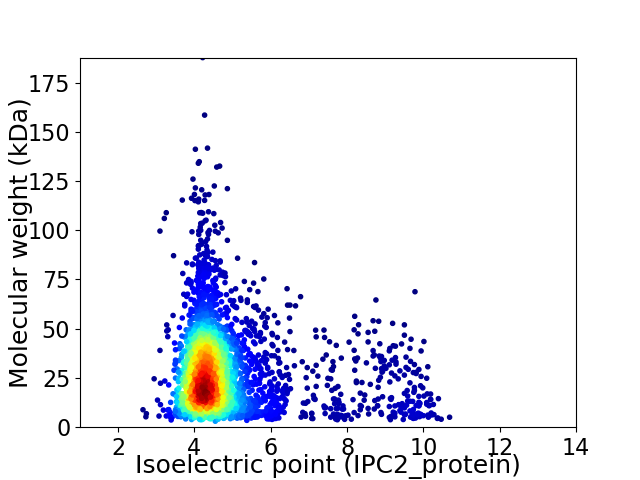
Virtual 2D-PAGE plot for 3340 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U5H185|A0A0U5H185_9EURY Homolog to tryptophan--tRNA ligase OS=Halobacterium hubeiense OX=1407499 GN=HHUB_1697 PE=4 SV=1
MM1 pKa = 7.13TRR3 pKa = 11.84DD4 pKa = 3.2VCVVVPTIRR13 pKa = 11.84EE14 pKa = 4.15YY15 pKa = 11.11EE16 pKa = 4.22CMRR19 pKa = 11.84AYY21 pKa = 9.73FQNARR26 pKa = 11.84DD27 pKa = 3.73HH28 pKa = 6.77GFDD31 pKa = 3.89LDD33 pKa = 3.75RR34 pKa = 11.84LFVVLVTEE42 pKa = 5.02DD43 pKa = 3.48FCDD46 pKa = 3.56TDD48 pKa = 3.74GMRR51 pKa = 11.84EE52 pKa = 3.9MLDD55 pKa = 3.45EE56 pKa = 4.71EE57 pKa = 5.08GVAGAVFDD65 pKa = 5.28GDD67 pKa = 6.02DD68 pKa = 3.28RR69 pKa = 11.84DD70 pKa = 3.44AWFDD74 pKa = 3.72EE75 pKa = 4.24QGLDD79 pKa = 3.83QYY81 pKa = 11.8SHH83 pKa = 7.69LIPAASHH90 pKa = 5.85AQTSFGLLYY99 pKa = 10.59LWANDD104 pKa = 3.23FEE106 pKa = 5.11YY107 pKa = 11.17GVFIDD112 pKa = 6.19DD113 pKa = 3.97DD114 pKa = 4.33TLPHH118 pKa = 7.13DD119 pKa = 3.54EE120 pKa = 4.0WDD122 pKa = 3.68FFGTHH127 pKa = 6.86LDD129 pKa = 3.7NLAHH133 pKa = 6.32EE134 pKa = 5.1GEE136 pKa = 4.39VEE138 pKa = 4.13EE139 pKa = 5.08VSSNEE144 pKa = 3.26QWVNVLYY151 pKa = 10.87QSDD154 pKa = 3.68SDD156 pKa = 4.29LYY158 pKa = 9.99PRR160 pKa = 11.84GYY162 pKa = 9.69PYY164 pKa = 10.2STMDD168 pKa = 3.93EE169 pKa = 4.4EE170 pKa = 5.38IEE172 pKa = 4.17TDD174 pKa = 3.36TTEE177 pKa = 3.98TDD179 pKa = 3.29HH180 pKa = 7.17VVASQGLWTNVPDD193 pKa = 4.12LDD195 pKa = 4.07AVRR198 pKa = 11.84ILMDD202 pKa = 4.92GDD204 pKa = 4.0LQGQAQTRR212 pKa = 11.84TDD214 pKa = 4.06FEE216 pKa = 5.05DD217 pKa = 3.77FDD219 pKa = 5.58RR220 pKa = 11.84DD221 pKa = 3.62FVAAEE226 pKa = 3.96DD227 pKa = 4.08NYY229 pKa = 10.86LTVCSMNLAFRR240 pKa = 11.84RR241 pKa = 11.84EE242 pKa = 4.16VVPAFYY248 pKa = 10.27QFPMDD253 pKa = 4.19DD254 pKa = 3.98NAWEE258 pKa = 4.08VGRR261 pKa = 11.84FDD263 pKa = 6.32DD264 pKa = 3.32IWSGVLLKK272 pKa = 10.61RR273 pKa = 11.84AADD276 pKa = 3.62VVGGEE281 pKa = 4.51IYY283 pKa = 10.83NGAPLCEE290 pKa = 4.47HH291 pKa = 6.51NKK293 pKa = 10.09APRR296 pKa = 11.84STFDD300 pKa = 3.55DD301 pKa = 4.0LANEE305 pKa = 4.35VAGLEE310 pKa = 4.27LNEE313 pKa = 4.58HH314 pKa = 5.91FWKK317 pKa = 10.56EE318 pKa = 3.97VAAVEE323 pKa = 4.1PDD325 pKa = 3.25EE326 pKa = 5.29DD327 pKa = 3.91SYY329 pKa = 11.86EE330 pKa = 4.05AVARR334 pKa = 11.84AVGEE338 pKa = 4.0RR339 pKa = 11.84LVEE342 pKa = 4.07SDD344 pKa = 3.62YY345 pKa = 11.89EE346 pKa = 4.29EE347 pKa = 4.58YY348 pKa = 11.42NNGAFLAEE356 pKa = 4.37CGEE359 pKa = 4.3YY360 pKa = 10.96LLDD363 pKa = 4.52WVDD366 pKa = 4.07CCAEE370 pKa = 4.04LAEE373 pKa = 4.14QRR375 pKa = 11.84AVAVADD381 pKa = 4.02DD382 pKa = 3.65
MM1 pKa = 7.13TRR3 pKa = 11.84DD4 pKa = 3.2VCVVVPTIRR13 pKa = 11.84EE14 pKa = 4.15YY15 pKa = 11.11EE16 pKa = 4.22CMRR19 pKa = 11.84AYY21 pKa = 9.73FQNARR26 pKa = 11.84DD27 pKa = 3.73HH28 pKa = 6.77GFDD31 pKa = 3.89LDD33 pKa = 3.75RR34 pKa = 11.84LFVVLVTEE42 pKa = 5.02DD43 pKa = 3.48FCDD46 pKa = 3.56TDD48 pKa = 3.74GMRR51 pKa = 11.84EE52 pKa = 3.9MLDD55 pKa = 3.45EE56 pKa = 4.71EE57 pKa = 5.08GVAGAVFDD65 pKa = 5.28GDD67 pKa = 6.02DD68 pKa = 3.28RR69 pKa = 11.84DD70 pKa = 3.44AWFDD74 pKa = 3.72EE75 pKa = 4.24QGLDD79 pKa = 3.83QYY81 pKa = 11.8SHH83 pKa = 7.69LIPAASHH90 pKa = 5.85AQTSFGLLYY99 pKa = 10.59LWANDD104 pKa = 3.23FEE106 pKa = 5.11YY107 pKa = 11.17GVFIDD112 pKa = 6.19DD113 pKa = 3.97DD114 pKa = 4.33TLPHH118 pKa = 7.13DD119 pKa = 3.54EE120 pKa = 4.0WDD122 pKa = 3.68FFGTHH127 pKa = 6.86LDD129 pKa = 3.7NLAHH133 pKa = 6.32EE134 pKa = 5.1GEE136 pKa = 4.39VEE138 pKa = 4.13EE139 pKa = 5.08VSSNEE144 pKa = 3.26QWVNVLYY151 pKa = 10.87QSDD154 pKa = 3.68SDD156 pKa = 4.29LYY158 pKa = 9.99PRR160 pKa = 11.84GYY162 pKa = 9.69PYY164 pKa = 10.2STMDD168 pKa = 3.93EE169 pKa = 4.4EE170 pKa = 5.38IEE172 pKa = 4.17TDD174 pKa = 3.36TTEE177 pKa = 3.98TDD179 pKa = 3.29HH180 pKa = 7.17VVASQGLWTNVPDD193 pKa = 4.12LDD195 pKa = 4.07AVRR198 pKa = 11.84ILMDD202 pKa = 4.92GDD204 pKa = 4.0LQGQAQTRR212 pKa = 11.84TDD214 pKa = 4.06FEE216 pKa = 5.05DD217 pKa = 3.77FDD219 pKa = 5.58RR220 pKa = 11.84DD221 pKa = 3.62FVAAEE226 pKa = 3.96DD227 pKa = 4.08NYY229 pKa = 10.86LTVCSMNLAFRR240 pKa = 11.84RR241 pKa = 11.84EE242 pKa = 4.16VVPAFYY248 pKa = 10.27QFPMDD253 pKa = 4.19DD254 pKa = 3.98NAWEE258 pKa = 4.08VGRR261 pKa = 11.84FDD263 pKa = 6.32DD264 pKa = 3.32IWSGVLLKK272 pKa = 10.61RR273 pKa = 11.84AADD276 pKa = 3.62VVGGEE281 pKa = 4.51IYY283 pKa = 10.83NGAPLCEE290 pKa = 4.47HH291 pKa = 6.51NKK293 pKa = 10.09APRR296 pKa = 11.84STFDD300 pKa = 3.55DD301 pKa = 4.0LANEE305 pKa = 4.35VAGLEE310 pKa = 4.27LNEE313 pKa = 4.58HH314 pKa = 5.91FWKK317 pKa = 10.56EE318 pKa = 3.97VAAVEE323 pKa = 4.1PDD325 pKa = 3.25EE326 pKa = 5.29DD327 pKa = 3.91SYY329 pKa = 11.86EE330 pKa = 4.05AVARR334 pKa = 11.84AVGEE338 pKa = 4.0RR339 pKa = 11.84LVEE342 pKa = 4.07SDD344 pKa = 3.62YY345 pKa = 11.89EE346 pKa = 4.29EE347 pKa = 4.58YY348 pKa = 11.42NNGAFLAEE356 pKa = 4.37CGEE359 pKa = 4.3YY360 pKa = 10.96LLDD363 pKa = 4.52WVDD366 pKa = 4.07CCAEE370 pKa = 4.04LAEE373 pKa = 4.14QRR375 pKa = 11.84AVAVADD381 pKa = 4.02DD382 pKa = 3.65
Molecular weight: 43.43 kDa
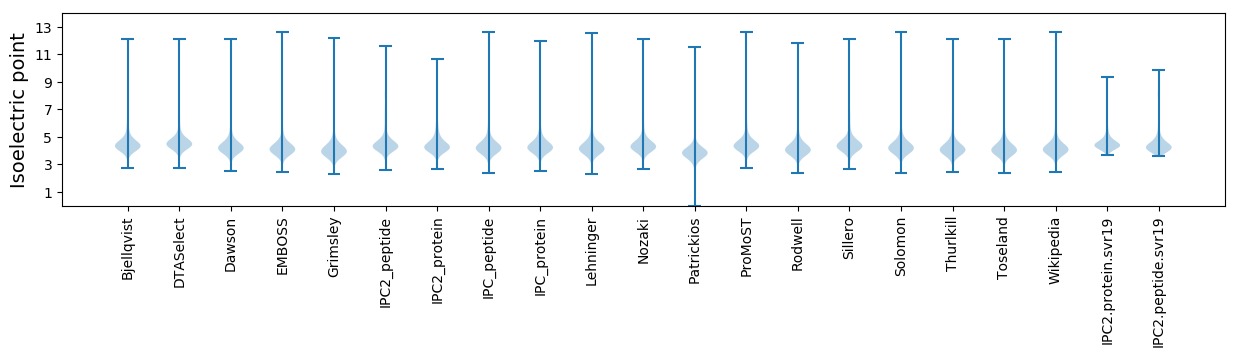
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U5AJY2|A0A0U5AJY2_9EURY Uncharacterized protein OS=Halobacterium hubeiense OX=1407499 GN=HHUB_4276 PE=4 SV=1
MM1 pKa = 7.63TEE3 pKa = 4.07YY4 pKa = 10.83QPGACNIGHH13 pKa = 6.43AEE15 pKa = 3.71RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 8.06LTGVAGFAAAALLVAAVATVHH41 pKa = 7.43ADD43 pKa = 4.13RR44 pKa = 11.84AWLLAAVAPLFVGFLGVLQARR65 pKa = 11.84ARR67 pKa = 11.84FCVGFAMAGVFDD79 pKa = 4.36VSASGDD85 pKa = 3.38RR86 pKa = 11.84RR87 pKa = 11.84QPAPEE92 pKa = 4.39AGQRR96 pKa = 11.84ADD98 pKa = 3.53RR99 pKa = 11.84KK100 pKa = 9.4RR101 pKa = 11.84AIVLSVKK108 pKa = 10.27ALAAATVGALALYY121 pKa = 9.65LAAGALAA128 pKa = 4.7
MM1 pKa = 7.63TEE3 pKa = 4.07YY4 pKa = 10.83QPGACNIGHH13 pKa = 6.43AEE15 pKa = 3.71RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 8.06LTGVAGFAAAALLVAAVATVHH41 pKa = 7.43ADD43 pKa = 4.13RR44 pKa = 11.84AWLLAAVAPLFVGFLGVLQARR65 pKa = 11.84ARR67 pKa = 11.84FCVGFAMAGVFDD79 pKa = 4.36VSASGDD85 pKa = 3.38RR86 pKa = 11.84RR87 pKa = 11.84QPAPEE92 pKa = 4.39AGQRR96 pKa = 11.84ADD98 pKa = 3.53RR99 pKa = 11.84KK100 pKa = 9.4RR101 pKa = 11.84AIVLSVKK108 pKa = 10.27ALAAATVGALALYY121 pKa = 9.65LAAGALAA128 pKa = 4.7
Molecular weight: 13.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
910421 |
28 |
1687 |
272.6 |
29.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.596 ± 0.081 | 0.677 ± 0.015 |
8.604 ± 0.063 | 8.541 ± 0.067 |
3.381 ± 0.029 | 8.325 ± 0.041 |
2.019 ± 0.023 | 3.432 ± 0.039 |
1.856 ± 0.027 | 8.849 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.685 ± 0.019 | 2.368 ± 0.027 |
4.552 ± 0.029 | 2.629 ± 0.027 |
6.312 ± 0.046 | 5.387 ± 0.034 |
6.192 ± 0.042 | 9.677 ± 0.056 |
1.147 ± 0.017 | 2.773 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
