
Desulfatibacillum aliphaticivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfatibacillum
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
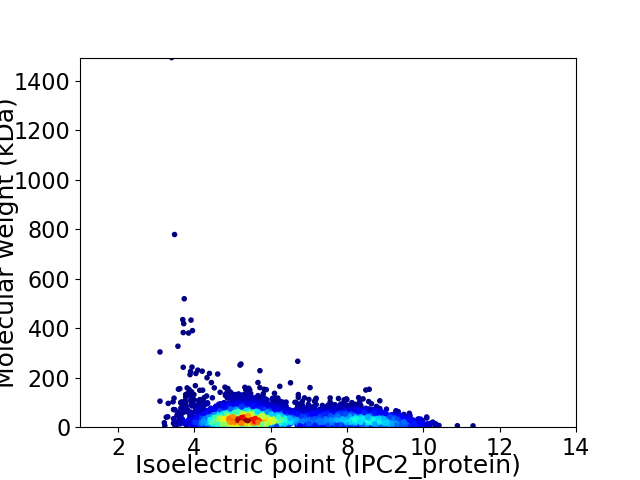
Virtual 2D-PAGE plot for 5198 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B8FAR4|B8FAR4_DESAL Methyltransferase type 12 OS=Desulfatibacillum aliphaticivorans OX=218208 GN=Dalk_1662 PE=4 SV=1
MM1 pKa = 7.63PCKK4 pKa = 10.43LYY6 pKa = 9.96WKK8 pKa = 10.21SFALSVLLLSFFIAFTGCSDD28 pKa = 4.83DD29 pKa = 6.22DD30 pKa = 4.03DD31 pKa = 4.33VLYY34 pKa = 11.2SGATSTGKK42 pKa = 10.4AVLGPLCGADD52 pKa = 3.32YY53 pKa = 11.1KK54 pKa = 11.39VFAYY58 pKa = 9.85PDD60 pKa = 3.62MKK62 pKa = 11.16NPIITGKK69 pKa = 9.14TSVSDD74 pKa = 4.26DD75 pKa = 3.49LSQAGLFSLPDD86 pKa = 3.46DD87 pKa = 4.2RR88 pKa = 11.84LADD91 pKa = 3.21GWLYY95 pKa = 10.69IMEE98 pKa = 4.48VTGGTDD104 pKa = 2.89VDD106 pKa = 4.17ADD108 pKa = 4.0DD109 pKa = 6.11DD110 pKa = 4.54GVIDD114 pKa = 5.44AAATPNLGTIHH125 pKa = 7.41LALQGEE131 pKa = 4.75TIKK134 pKa = 11.11KK135 pKa = 9.55EE136 pKa = 3.98GFRR139 pKa = 11.84VNILSEE145 pKa = 4.3LVYY148 pKa = 9.57QTVSFMLMAGYY159 pKa = 8.29PQSDD163 pKa = 3.86VIARR167 pKa = 11.84MNAAASRR174 pKa = 11.84LINQDD179 pKa = 2.75LDD181 pKa = 3.9GNGQIDD187 pKa = 4.38HH188 pKa = 7.5KK189 pKa = 11.21DD190 pKa = 3.16LAYY193 pKa = 9.7WEE195 pKa = 4.41PVNNRR200 pKa = 11.84GALVRR205 pKa = 11.84GWASITPCIDD215 pKa = 3.04AVHH218 pKa = 6.52NNDD221 pKa = 4.81DD222 pKa = 4.17LFSLAVNRR230 pKa = 11.84TSPQSALANTPGTALNVAVDD250 pKa = 3.47GGFAYY255 pKa = 10.62VADD258 pKa = 4.5ADD260 pKa = 4.27GGLHH264 pKa = 6.52VFDD267 pKa = 4.39TTDD270 pKa = 3.26PFDD273 pKa = 4.16PVLLGTAAMDD283 pKa = 3.94RR284 pKa = 11.84ARR286 pKa = 11.84DD287 pKa = 3.76VVISGTYY294 pKa = 10.19AYY296 pKa = 10.61VADD299 pKa = 4.56DD300 pKa = 4.2AEE302 pKa = 4.32GLKK305 pKa = 10.17IVDD308 pKa = 4.26AADD311 pKa = 3.89PSDD314 pKa = 3.77PQVVGVVDD322 pKa = 3.7TEE324 pKa = 3.95KK325 pKa = 10.99ARR327 pKa = 11.84AVVVRR332 pKa = 11.84GNYY335 pKa = 10.34AYY337 pKa = 10.89VADD340 pKa = 4.78DD341 pKa = 4.03ANGLRR346 pKa = 11.84VIDD349 pKa = 4.02VADD352 pKa = 3.89PANPQIVGTAPTVKK366 pKa = 10.55ANDD369 pKa = 3.14VALFGDD375 pKa = 4.49YY376 pKa = 11.06ACVADD381 pKa = 4.78DD382 pKa = 3.63VGGVKK387 pKa = 10.17VVDD390 pKa = 3.55IADD393 pKa = 3.94PANPQVVGTAATEE406 pKa = 4.26DD407 pKa = 3.6ASSAAIADD415 pKa = 4.56GYY417 pKa = 11.64AYY419 pKa = 10.74VADD422 pKa = 3.74SAFGIKK428 pKa = 10.03VVDD431 pKa = 4.78LSDD434 pKa = 3.24PTAPVVVGAASTGFSRR450 pKa = 11.84SMEE453 pKa = 3.97VFNRR457 pKa = 11.84VAYY460 pKa = 8.63VTGDD464 pKa = 3.04SDD466 pKa = 3.82GFKK469 pKa = 10.13MVDD472 pKa = 3.05VSDD475 pKa = 4.01PANPKK480 pKa = 10.21IIGTRR485 pKa = 11.84DD486 pKa = 3.07EE487 pKa = 4.63GDD489 pKa = 3.34TRR491 pKa = 11.84GLWIGDD497 pKa = 3.72NIAYY501 pKa = 8.92LANGYY506 pKa = 9.67QGLKK510 pKa = 8.55VTDD513 pKa = 3.27ITYY516 pKa = 10.16EE517 pKa = 3.98PNPQVIGCVNTADD530 pKa = 3.82AQDD533 pKa = 3.45AAFQGDD539 pKa = 4.04YY540 pKa = 10.97AYY542 pKa = 11.02VADD545 pKa = 4.52GAGGLAVVDD554 pKa = 4.31ISDD557 pKa = 3.91PANPVLIGSAPSTFAQGAAVQGDD580 pKa = 4.04YY581 pKa = 11.05AYY583 pKa = 11.03VADD586 pKa = 4.96GGDD589 pKa = 3.49GLRR592 pKa = 11.84IFDD595 pKa = 4.18VSDD598 pKa = 3.43PANPFEE604 pKa = 4.25TSVLDD609 pKa = 3.6TDD611 pKa = 4.32DD612 pKa = 4.49ANIVAVSGDD621 pKa = 3.66YY622 pKa = 10.82AYY624 pKa = 11.1VADD627 pKa = 5.04GSAGLRR633 pKa = 11.84VIDD636 pKa = 3.8VSNPYY641 pKa = 10.22SPQLKK646 pKa = 8.47GTLDD650 pKa = 3.17TRR652 pKa = 11.84SAYY655 pKa = 10.36DD656 pKa = 3.33VTVSGDD662 pKa = 3.22LAYY665 pKa = 11.06VGDD668 pKa = 4.5GMDD671 pKa = 3.53GLIIVDD677 pKa = 3.68VSDD680 pKa = 4.64PEE682 pKa = 4.17NPDD685 pKa = 4.18DD686 pKa = 5.58IGVLDD691 pKa = 4.38TFFADD696 pKa = 6.05DD697 pKa = 3.75IVLDD701 pKa = 3.86GNYY704 pKa = 10.39AYY706 pKa = 9.79IADD709 pKa = 4.57GPQGLKK715 pKa = 10.31VADD718 pKa = 4.04ISDD721 pKa = 3.77PTAPEE726 pKa = 3.44QKK728 pKa = 10.59AYY730 pKa = 11.06LDD732 pKa = 3.97TPNSCRR738 pKa = 11.84ALALSGNYY746 pKa = 9.86LYY748 pKa = 10.98AADD751 pKa = 5.11NGPDD755 pKa = 3.67MIIVDD760 pKa = 4.12VSDD763 pKa = 3.9PLNPVEE769 pKa = 4.23IAWVEE774 pKa = 4.09TLNVFYY780 pKa = 10.57RR781 pKa = 11.84LEE783 pKa = 3.89VHH785 pKa = 6.77EE786 pKa = 5.43GYY788 pKa = 10.86LYY790 pKa = 9.77LTSSEE795 pKa = 5.07LGLQVLKK802 pKa = 10.81AVPAPP807 pKa = 3.64
MM1 pKa = 7.63PCKK4 pKa = 10.43LYY6 pKa = 9.96WKK8 pKa = 10.21SFALSVLLLSFFIAFTGCSDD28 pKa = 4.83DD29 pKa = 6.22DD30 pKa = 4.03DD31 pKa = 4.33VLYY34 pKa = 11.2SGATSTGKK42 pKa = 10.4AVLGPLCGADD52 pKa = 3.32YY53 pKa = 11.1KK54 pKa = 11.39VFAYY58 pKa = 9.85PDD60 pKa = 3.62MKK62 pKa = 11.16NPIITGKK69 pKa = 9.14TSVSDD74 pKa = 4.26DD75 pKa = 3.49LSQAGLFSLPDD86 pKa = 3.46DD87 pKa = 4.2RR88 pKa = 11.84LADD91 pKa = 3.21GWLYY95 pKa = 10.69IMEE98 pKa = 4.48VTGGTDD104 pKa = 2.89VDD106 pKa = 4.17ADD108 pKa = 4.0DD109 pKa = 6.11DD110 pKa = 4.54GVIDD114 pKa = 5.44AAATPNLGTIHH125 pKa = 7.41LALQGEE131 pKa = 4.75TIKK134 pKa = 11.11KK135 pKa = 9.55EE136 pKa = 3.98GFRR139 pKa = 11.84VNILSEE145 pKa = 4.3LVYY148 pKa = 9.57QTVSFMLMAGYY159 pKa = 8.29PQSDD163 pKa = 3.86VIARR167 pKa = 11.84MNAAASRR174 pKa = 11.84LINQDD179 pKa = 2.75LDD181 pKa = 3.9GNGQIDD187 pKa = 4.38HH188 pKa = 7.5KK189 pKa = 11.21DD190 pKa = 3.16LAYY193 pKa = 9.7WEE195 pKa = 4.41PVNNRR200 pKa = 11.84GALVRR205 pKa = 11.84GWASITPCIDD215 pKa = 3.04AVHH218 pKa = 6.52NNDD221 pKa = 4.81DD222 pKa = 4.17LFSLAVNRR230 pKa = 11.84TSPQSALANTPGTALNVAVDD250 pKa = 3.47GGFAYY255 pKa = 10.62VADD258 pKa = 4.5ADD260 pKa = 4.27GGLHH264 pKa = 6.52VFDD267 pKa = 4.39TTDD270 pKa = 3.26PFDD273 pKa = 4.16PVLLGTAAMDD283 pKa = 3.94RR284 pKa = 11.84ARR286 pKa = 11.84DD287 pKa = 3.76VVISGTYY294 pKa = 10.19AYY296 pKa = 10.61VADD299 pKa = 4.56DD300 pKa = 4.2AEE302 pKa = 4.32GLKK305 pKa = 10.17IVDD308 pKa = 4.26AADD311 pKa = 3.89PSDD314 pKa = 3.77PQVVGVVDD322 pKa = 3.7TEE324 pKa = 3.95KK325 pKa = 10.99ARR327 pKa = 11.84AVVVRR332 pKa = 11.84GNYY335 pKa = 10.34AYY337 pKa = 10.89VADD340 pKa = 4.78DD341 pKa = 4.03ANGLRR346 pKa = 11.84VIDD349 pKa = 4.02VADD352 pKa = 3.89PANPQIVGTAPTVKK366 pKa = 10.55ANDD369 pKa = 3.14VALFGDD375 pKa = 4.49YY376 pKa = 11.06ACVADD381 pKa = 4.78DD382 pKa = 3.63VGGVKK387 pKa = 10.17VVDD390 pKa = 3.55IADD393 pKa = 3.94PANPQVVGTAATEE406 pKa = 4.26DD407 pKa = 3.6ASSAAIADD415 pKa = 4.56GYY417 pKa = 11.64AYY419 pKa = 10.74VADD422 pKa = 3.74SAFGIKK428 pKa = 10.03VVDD431 pKa = 4.78LSDD434 pKa = 3.24PTAPVVVGAASTGFSRR450 pKa = 11.84SMEE453 pKa = 3.97VFNRR457 pKa = 11.84VAYY460 pKa = 8.63VTGDD464 pKa = 3.04SDD466 pKa = 3.82GFKK469 pKa = 10.13MVDD472 pKa = 3.05VSDD475 pKa = 4.01PANPKK480 pKa = 10.21IIGTRR485 pKa = 11.84DD486 pKa = 3.07EE487 pKa = 4.63GDD489 pKa = 3.34TRR491 pKa = 11.84GLWIGDD497 pKa = 3.72NIAYY501 pKa = 8.92LANGYY506 pKa = 9.67QGLKK510 pKa = 8.55VTDD513 pKa = 3.27ITYY516 pKa = 10.16EE517 pKa = 3.98PNPQVIGCVNTADD530 pKa = 3.82AQDD533 pKa = 3.45AAFQGDD539 pKa = 4.04YY540 pKa = 10.97AYY542 pKa = 11.02VADD545 pKa = 4.52GAGGLAVVDD554 pKa = 4.31ISDD557 pKa = 3.91PANPVLIGSAPSTFAQGAAVQGDD580 pKa = 4.04YY581 pKa = 11.05AYY583 pKa = 11.03VADD586 pKa = 4.96GGDD589 pKa = 3.49GLRR592 pKa = 11.84IFDD595 pKa = 4.18VSDD598 pKa = 3.43PANPFEE604 pKa = 4.25TSVLDD609 pKa = 3.6TDD611 pKa = 4.32DD612 pKa = 4.49ANIVAVSGDD621 pKa = 3.66YY622 pKa = 10.82AYY624 pKa = 11.1VADD627 pKa = 5.04GSAGLRR633 pKa = 11.84VIDD636 pKa = 3.8VSNPYY641 pKa = 10.22SPQLKK646 pKa = 8.47GTLDD650 pKa = 3.17TRR652 pKa = 11.84SAYY655 pKa = 10.36DD656 pKa = 3.33VTVSGDD662 pKa = 3.22LAYY665 pKa = 11.06VGDD668 pKa = 4.5GMDD671 pKa = 3.53GLIIVDD677 pKa = 3.68VSDD680 pKa = 4.64PEE682 pKa = 4.17NPDD685 pKa = 4.18DD686 pKa = 5.58IGVLDD691 pKa = 4.38TFFADD696 pKa = 6.05DD697 pKa = 3.75IVLDD701 pKa = 3.86GNYY704 pKa = 10.39AYY706 pKa = 9.79IADD709 pKa = 4.57GPQGLKK715 pKa = 10.31VADD718 pKa = 4.04ISDD721 pKa = 3.77PTAPEE726 pKa = 3.44QKK728 pKa = 10.59AYY730 pKa = 11.06LDD732 pKa = 3.97TPNSCRR738 pKa = 11.84ALALSGNYY746 pKa = 9.86LYY748 pKa = 10.98AADD751 pKa = 5.11NGPDD755 pKa = 3.67MIIVDD760 pKa = 4.12VSDD763 pKa = 3.9PLNPVEE769 pKa = 4.23IAWVEE774 pKa = 4.09TLNVFYY780 pKa = 10.57RR781 pKa = 11.84LEE783 pKa = 3.89VHH785 pKa = 6.77EE786 pKa = 5.43GYY788 pKa = 10.86LYY790 pKa = 9.77LTSSEE795 pKa = 5.07LGLQVLKK802 pKa = 10.81AVPAPP807 pKa = 3.64
Molecular weight: 84.53 kDa
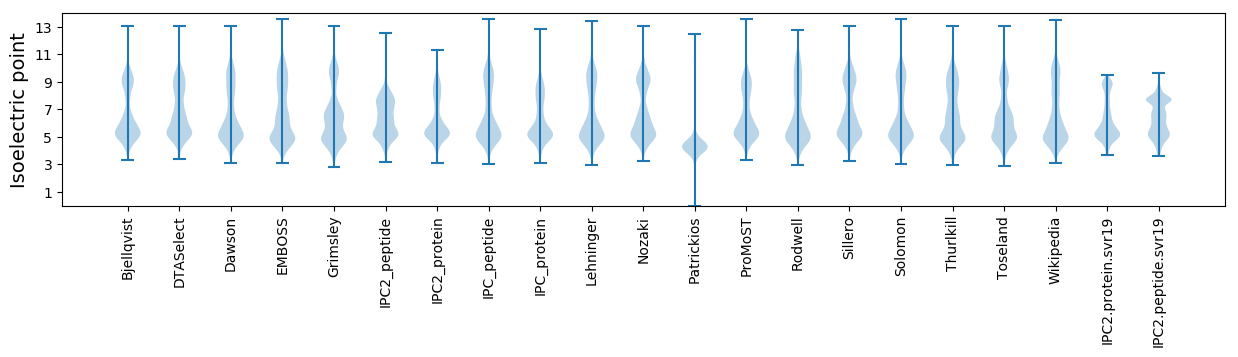
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B8FN68|B8FN68_DESAL Ion-translocating oxidoreductase complex subunit A OS=Desulfatibacillum aliphaticivorans OX=218208 GN=rnfA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.24IKK11 pKa = 10.37RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.37GFLKK20 pKa = 10.63RR21 pKa = 11.84MSTKK25 pKa = 10.08AGRR28 pKa = 11.84RR29 pKa = 11.84IIKK32 pKa = 9.74RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.98RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.24IKK11 pKa = 10.37RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 6.37GFLKK20 pKa = 10.63RR21 pKa = 11.84MSTKK25 pKa = 10.08AGRR28 pKa = 11.84RR29 pKa = 11.84IIKK32 pKa = 9.74RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.98RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1868736 |
31 |
15349 |
359.5 |
39.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.395 ± 0.05 | 1.331 ± 0.017 |
5.866 ± 0.048 | 6.594 ± 0.048 |
4.203 ± 0.034 | 7.969 ± 0.065 |
1.889 ± 0.017 | 5.917 ± 0.03 |
5.644 ± 0.057 | 9.571 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.97 ± 0.024 | 3.65 ± 0.033 |
4.612 ± 0.034 | 3.417 ± 0.02 |
4.971 ± 0.043 | 5.829 ± 0.038 |
4.941 ± 0.064 | 7.03 ± 0.037 |
1.194 ± 0.014 | 3.007 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
