
Beta vulgaris subsp. vulgaris (Beet)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales;
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
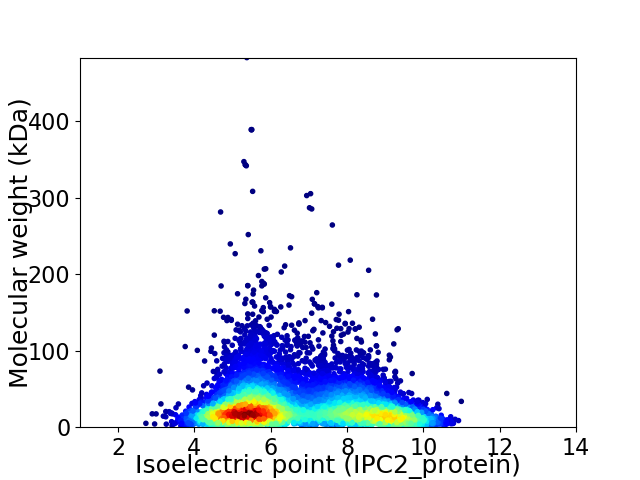
Virtual 2D-PAGE plot for 7611 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
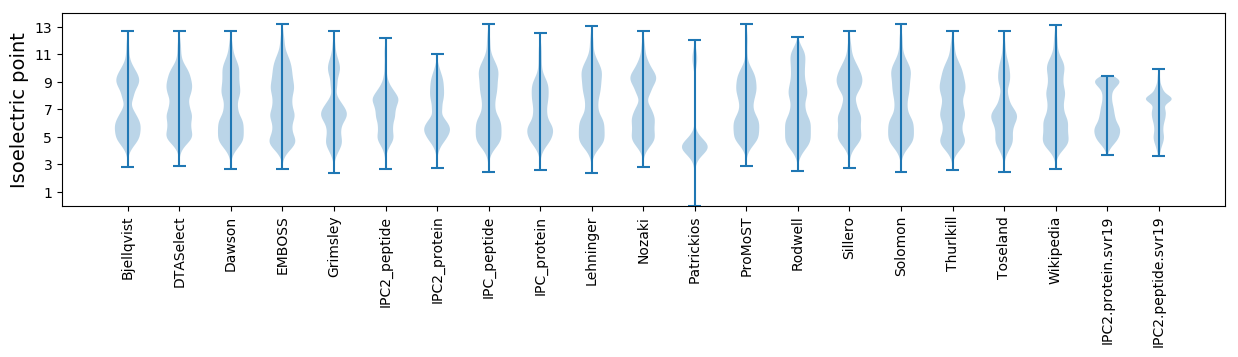
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0J8DSF5|A0A0J8DSF5_BETVV Uncharacterized protein OS=Beta vulgaris subsp. vulgaris OX=3555 GN=BVRB_029180 PE=4 SV=1
MM1 pKa = 7.29NKK3 pKa = 9.91SLALALGALMVASPALAQEE22 pKa = 4.12EE23 pKa = 4.47AVLNVYY29 pKa = 9.34NWSDD33 pKa = 4.06YY34 pKa = 10.51IAEE37 pKa = 3.92DD38 pKa = 3.95TIANFEE44 pKa = 4.08AATGIKK50 pKa = 10.12VNYY53 pKa = 9.54DD54 pKa = 3.15VYY56 pKa = 11.54DD57 pKa = 3.63NNEE60 pKa = 3.78IVDD63 pKa = 4.06AKK65 pKa = 10.85LLAGSSGYY73 pKa = 10.79DD74 pKa = 2.68IVVPSGNFLEE84 pKa = 4.48RR85 pKa = 11.84QIQAGLILPLDD96 pKa = 3.78KK97 pKa = 11.27SKK99 pKa = 9.84LTNLGNLDD107 pKa = 4.01PAVMATAAAQDD118 pKa = 3.86PDD120 pKa = 3.72NAHH123 pKa = 6.26GVPYY127 pKa = 9.78MINTIGYY134 pKa = 8.36GYY136 pKa = 10.71NVAKK140 pKa = 9.42VTEE143 pKa = 4.21ILGADD148 pKa = 3.73APVDD152 pKa = 3.35SWDD155 pKa = 5.1LIFKK159 pKa = 10.5PEE161 pKa = 3.85FAEE164 pKa = 4.25KK165 pKa = 10.56LSACGISLLDD175 pKa = 3.71SPSEE179 pKa = 4.09VMGIALNYY187 pKa = 10.26LGLDD191 pKa = 3.36ANSEE195 pKa = 4.24SEE197 pKa = 4.04EE198 pKa = 4.07DD199 pKa = 3.33LAKK202 pKa = 10.78AEE204 pKa = 4.11EE205 pKa = 5.17LINAIKK211 pKa = 10.48PFIRR215 pKa = 11.84YY216 pKa = 7.94FNSSQYY222 pKa = 10.49IDD224 pKa = 4.31DD225 pKa = 4.69LGNGEE230 pKa = 4.2TCVALGYY237 pKa = 10.59SGDD240 pKa = 3.41IFIAADD246 pKa = 3.15AA247 pKa = 4.59
MM1 pKa = 7.29NKK3 pKa = 9.91SLALALGALMVASPALAQEE22 pKa = 4.12EE23 pKa = 4.47AVLNVYY29 pKa = 9.34NWSDD33 pKa = 4.06YY34 pKa = 10.51IAEE37 pKa = 3.92DD38 pKa = 3.95TIANFEE44 pKa = 4.08AATGIKK50 pKa = 10.12VNYY53 pKa = 9.54DD54 pKa = 3.15VYY56 pKa = 11.54DD57 pKa = 3.63NNEE60 pKa = 3.78IVDD63 pKa = 4.06AKK65 pKa = 10.85LLAGSSGYY73 pKa = 10.79DD74 pKa = 2.68IVVPSGNFLEE84 pKa = 4.48RR85 pKa = 11.84QIQAGLILPLDD96 pKa = 3.78KK97 pKa = 11.27SKK99 pKa = 9.84LTNLGNLDD107 pKa = 4.01PAVMATAAAQDD118 pKa = 3.86PDD120 pKa = 3.72NAHH123 pKa = 6.26GVPYY127 pKa = 9.78MINTIGYY134 pKa = 8.36GYY136 pKa = 10.71NVAKK140 pKa = 9.42VTEE143 pKa = 4.21ILGADD148 pKa = 3.73APVDD152 pKa = 3.35SWDD155 pKa = 5.1LIFKK159 pKa = 10.5PEE161 pKa = 3.85FAEE164 pKa = 4.25KK165 pKa = 10.56LSACGISLLDD175 pKa = 3.71SPSEE179 pKa = 4.09VMGIALNYY187 pKa = 10.26LGLDD191 pKa = 3.36ANSEE195 pKa = 4.24SEE197 pKa = 4.04EE198 pKa = 4.07DD199 pKa = 3.33LAKK202 pKa = 10.78AEE204 pKa = 4.11EE205 pKa = 5.17LINAIKK211 pKa = 10.48PFIRR215 pKa = 11.84YY216 pKa = 7.94FNSSQYY222 pKa = 10.49IDD224 pKa = 4.31DD225 pKa = 4.69LGNGEE230 pKa = 4.2TCVALGYY237 pKa = 10.59SGDD240 pKa = 3.41IFIAADD246 pKa = 3.15AA247 pKa = 4.59
Molecular weight: 26.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0J7YMS7|A0A0J7YMS7_BETVV Serine/threonine-protein phosphatase 2A activator (Fragment) OS=Beta vulgaris subsp. vulgaris OX=3555 GN=BVRB_041550 PE=3 SV=1
SS1 pKa = 6.84TIFTARR7 pKa = 11.84RR8 pKa = 11.84PDD10 pKa = 3.03SMLQRR15 pKa = 11.84RR16 pKa = 11.84IFTARR21 pKa = 11.84RR22 pKa = 11.84PFVRR26 pKa = 11.84RR27 pKa = 11.84SNRR30 pKa = 11.84LRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84SSRR37 pKa = 11.84LRR39 pKa = 11.84RR40 pKa = 11.84FFRR43 pKa = 11.84WSFQQATSSFRR54 pKa = 11.84SFSFQQAAAEE64 pKa = 4.14FRR66 pKa = 11.84SSFVGG71 pKa = 3.44
SS1 pKa = 6.84TIFTARR7 pKa = 11.84RR8 pKa = 11.84PDD10 pKa = 3.03SMLQRR15 pKa = 11.84RR16 pKa = 11.84IFTARR21 pKa = 11.84RR22 pKa = 11.84PFVRR26 pKa = 11.84RR27 pKa = 11.84SNRR30 pKa = 11.84LRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84SSRR37 pKa = 11.84LRR39 pKa = 11.84RR40 pKa = 11.84FFRR43 pKa = 11.84WSFQQATSSFRR54 pKa = 11.84SFSFQQAAAEE64 pKa = 4.14FRR66 pKa = 11.84SSFVGG71 pKa = 3.44
Molecular weight: 8.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2194387 |
8 |
4316 |
288.3 |
32.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.921 ± 0.033 | 1.871 ± 0.016 |
5.514 ± 0.027 | 6.307 ± 0.034 |
4.049 ± 0.021 | 6.254 ± 0.043 |
2.441 ± 0.017 | 5.387 ± 0.026 |
5.893 ± 0.029 | 9.634 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.439 ± 0.012 | 4.479 ± 0.023 |
4.963 ± 0.035 | 3.892 ± 0.027 |
5.461 ± 0.029 | 8.986 ± 0.038 |
4.985 ± 0.02 | 6.541 ± 0.023 |
1.215 ± 0.013 | 2.767 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
