
Digitaria streak virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
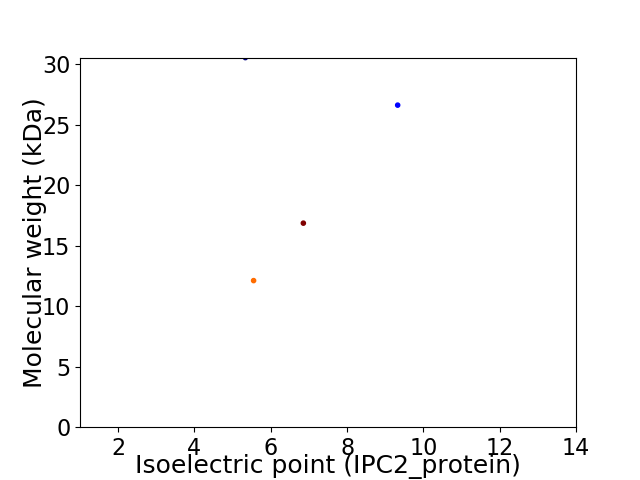
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
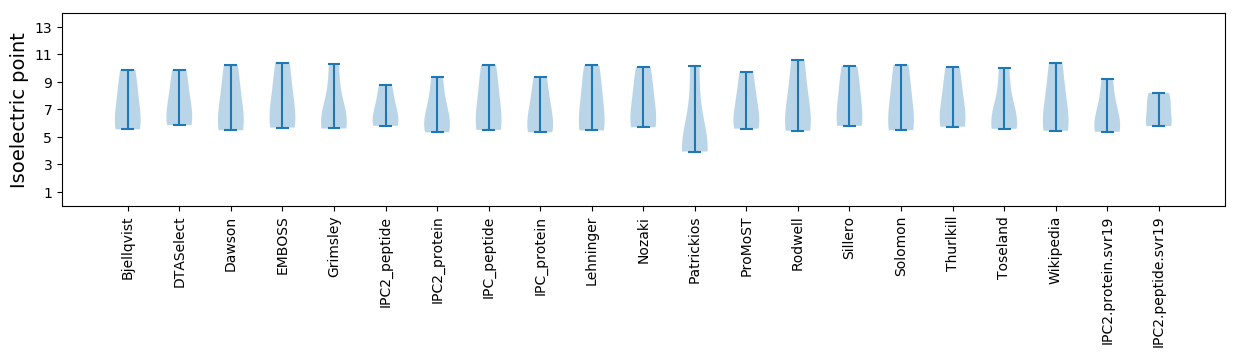
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q67568|Q67568_9GEMI Replication-associated protein OS=Digitaria streak virus OX=10837 PE=3 SV=1
MM1 pKa = 7.4AANRR5 pKa = 11.84SFRR8 pKa = 11.84HH9 pKa = 5.38RR10 pKa = 11.84NANTFLTYY18 pKa = 10.33SKK20 pKa = 10.32CDD22 pKa = 3.45HH23 pKa = 6.64SPQLIADD30 pKa = 4.38HH31 pKa = 6.87LWDD34 pKa = 5.57LLKK37 pKa = 10.61SWNPIYY43 pKa = 10.48ILVASEE49 pKa = 3.78HH50 pKa = 6.43HH51 pKa = 6.98ADD53 pKa = 3.99GSLHH57 pKa = 5.62SHH59 pKa = 6.91ALVQTEE65 pKa = 4.2KK66 pKa = 10.77QVNTTNQRR74 pKa = 11.84FFDD77 pKa = 3.28ILEE80 pKa = 3.86FHH82 pKa = 7.37PNIQSAKK89 pKa = 9.09SVNKK93 pKa = 9.47VRR95 pKa = 11.84TYY97 pKa = 10.19ILKK100 pKa = 10.23NPVEE104 pKa = 4.16KK105 pKa = 10.6FEE107 pKa = 5.15RR108 pKa = 11.84GTFVPRR114 pKa = 11.84KK115 pKa = 9.73SPFLGEE121 pKa = 4.18SSSSEE126 pKa = 3.9KK127 pKa = 10.51KK128 pKa = 9.9HH129 pKa = 6.58NKK131 pKa = 9.2DD132 pKa = 3.28DD133 pKa = 3.69VMRR136 pKa = 11.84DD137 pKa = 3.6IIDD140 pKa = 3.68HH141 pKa = 5.73ATSKK145 pKa = 11.07EE146 pKa = 4.22EE147 pKa = 3.96YY148 pKa = 10.48LSMVQKK154 pKa = 10.83ALPYY158 pKa = 10.55DD159 pKa = 3.43WATKK163 pKa = 10.42LSYY166 pKa = 10.68FEE168 pKa = 4.67YY169 pKa = 10.61SADD172 pKa = 3.43RR173 pKa = 11.84LFPVEE178 pKa = 4.48AAPFINPHH186 pKa = 6.56PPSEE190 pKa = 4.54PDD192 pKa = 3.58LLCQEE197 pKa = 5.49TITDD201 pKa = 3.79WLQNDD206 pKa = 4.6LFQVSAEE213 pKa = 4.37AYY215 pKa = 9.48CLLNPTCYY223 pKa = 9.7TRR225 pKa = 11.84EE226 pKa = 3.8EE227 pKa = 4.77AISDD231 pKa = 4.17LQWMSDD237 pKa = 3.63YY238 pKa = 10.39TRR240 pKa = 11.84SRR242 pKa = 11.84MGSEE246 pKa = 3.85NAASTSSVQQEE257 pKa = 4.44LEE259 pKa = 4.14SLLGPEE265 pKa = 4.28AA266 pKa = 4.5
MM1 pKa = 7.4AANRR5 pKa = 11.84SFRR8 pKa = 11.84HH9 pKa = 5.38RR10 pKa = 11.84NANTFLTYY18 pKa = 10.33SKK20 pKa = 10.32CDD22 pKa = 3.45HH23 pKa = 6.64SPQLIADD30 pKa = 4.38HH31 pKa = 6.87LWDD34 pKa = 5.57LLKK37 pKa = 10.61SWNPIYY43 pKa = 10.48ILVASEE49 pKa = 3.78HH50 pKa = 6.43HH51 pKa = 6.98ADD53 pKa = 3.99GSLHH57 pKa = 5.62SHH59 pKa = 6.91ALVQTEE65 pKa = 4.2KK66 pKa = 10.77QVNTTNQRR74 pKa = 11.84FFDD77 pKa = 3.28ILEE80 pKa = 3.86FHH82 pKa = 7.37PNIQSAKK89 pKa = 9.09SVNKK93 pKa = 9.47VRR95 pKa = 11.84TYY97 pKa = 10.19ILKK100 pKa = 10.23NPVEE104 pKa = 4.16KK105 pKa = 10.6FEE107 pKa = 5.15RR108 pKa = 11.84GTFVPRR114 pKa = 11.84KK115 pKa = 9.73SPFLGEE121 pKa = 4.18SSSSEE126 pKa = 3.9KK127 pKa = 10.51KK128 pKa = 9.9HH129 pKa = 6.58NKK131 pKa = 9.2DD132 pKa = 3.28DD133 pKa = 3.69VMRR136 pKa = 11.84DD137 pKa = 3.6IIDD140 pKa = 3.68HH141 pKa = 5.73ATSKK145 pKa = 11.07EE146 pKa = 4.22EE147 pKa = 3.96YY148 pKa = 10.48LSMVQKK154 pKa = 10.83ALPYY158 pKa = 10.55DD159 pKa = 3.43WATKK163 pKa = 10.42LSYY166 pKa = 10.68FEE168 pKa = 4.67YY169 pKa = 10.61SADD172 pKa = 3.43RR173 pKa = 11.84LFPVEE178 pKa = 4.48AAPFINPHH186 pKa = 6.56PPSEE190 pKa = 4.54PDD192 pKa = 3.58LLCQEE197 pKa = 5.49TITDD201 pKa = 3.79WLQNDD206 pKa = 4.6LFQVSAEE213 pKa = 4.37AYY215 pKa = 9.48CLLNPTCYY223 pKa = 9.7TRR225 pKa = 11.84EE226 pKa = 3.8EE227 pKa = 4.77AISDD231 pKa = 4.17LQWMSDD237 pKa = 3.63YY238 pKa = 10.39TRR240 pKa = 11.84SRR242 pKa = 11.84MGSEE246 pKa = 3.85NAASTSSVQQEE257 pKa = 4.44LEE259 pKa = 4.14SLLGPEE265 pKa = 4.28AA266 pKa = 4.5
Molecular weight: 30.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q67567|Q67567_9GEMI Uncharacterized protein (Fragment) OS=Digitaria streak virus OX=10837 PE=4 SV=1
MM1 pKa = 7.26SSSMKK6 pKa = 10.25RR7 pKa = 11.84KK8 pKa = 8.92RR9 pKa = 11.84TDD11 pKa = 2.87EE12 pKa = 4.32GSGSRR17 pKa = 11.84RR18 pKa = 11.84FTKK21 pKa = 10.25KK22 pKa = 9.81KK23 pKa = 9.65SAAKK27 pKa = 10.19GRR29 pKa = 11.84TSSSRR34 pKa = 11.84AIRR37 pKa = 11.84PALQIQSFVSAGAATIAVPTGGVCHH62 pKa = 7.27LLSSYY67 pKa = 11.26SRR69 pKa = 11.84GSGEE73 pKa = 4.1GDD75 pKa = 2.58RR76 pKa = 11.84HH77 pKa = 5.17TNEE80 pKa = 3.68TVTYY84 pKa = 9.43KK85 pKa = 10.89AAFDD89 pKa = 3.69YY90 pKa = 10.99HH91 pKa = 7.06FSANAGPCAYY101 pKa = 10.34SSIGVGVLWLVYY113 pKa = 9.84DD114 pKa = 4.52AQPSGQVPAVTDD126 pKa = 3.76IFPHH130 pKa = 6.35DD131 pKa = 3.64TTLQSFPYY139 pKa = 7.3TWKK142 pKa = 10.2VGRR145 pKa = 11.84EE146 pKa = 3.83VCHH149 pKa = 6.21RR150 pKa = 11.84FVVKK154 pKa = 10.44RR155 pKa = 11.84RR156 pKa = 11.84WCFTMEE162 pKa = 3.97TNGRR166 pKa = 11.84IGSDD170 pKa = 3.39KK171 pKa = 10.7PPSNAVWPPCKK182 pKa = 9.78RR183 pKa = 11.84SIYY186 pKa = 8.45FHH188 pKa = 7.11KK189 pKa = 10.18FATGLGVKK197 pKa = 7.95TEE199 pKa = 4.21WKK201 pKa = 8.12NTTDD205 pKa = 3.62GGVGSIKK212 pKa = 10.37KK213 pKa = 9.07GALYY217 pKa = 9.47FVIAPGSGIDD227 pKa = 3.5FTLFGTCRR235 pKa = 11.84MYY237 pKa = 10.82FKK239 pKa = 10.94SVGNQQ244 pKa = 2.8
MM1 pKa = 7.26SSSMKK6 pKa = 10.25RR7 pKa = 11.84KK8 pKa = 8.92RR9 pKa = 11.84TDD11 pKa = 2.87EE12 pKa = 4.32GSGSRR17 pKa = 11.84RR18 pKa = 11.84FTKK21 pKa = 10.25KK22 pKa = 9.81KK23 pKa = 9.65SAAKK27 pKa = 10.19GRR29 pKa = 11.84TSSSRR34 pKa = 11.84AIRR37 pKa = 11.84PALQIQSFVSAGAATIAVPTGGVCHH62 pKa = 7.27LLSSYY67 pKa = 11.26SRR69 pKa = 11.84GSGEE73 pKa = 4.1GDD75 pKa = 2.58RR76 pKa = 11.84HH77 pKa = 5.17TNEE80 pKa = 3.68TVTYY84 pKa = 9.43KK85 pKa = 10.89AAFDD89 pKa = 3.69YY90 pKa = 10.99HH91 pKa = 7.06FSANAGPCAYY101 pKa = 10.34SSIGVGVLWLVYY113 pKa = 9.84DD114 pKa = 4.52AQPSGQVPAVTDD126 pKa = 3.76IFPHH130 pKa = 6.35DD131 pKa = 3.64TTLQSFPYY139 pKa = 7.3TWKK142 pKa = 10.2VGRR145 pKa = 11.84EE146 pKa = 3.83VCHH149 pKa = 6.21RR150 pKa = 11.84FVVKK154 pKa = 10.44RR155 pKa = 11.84RR156 pKa = 11.84WCFTMEE162 pKa = 3.97TNGRR166 pKa = 11.84IGSDD170 pKa = 3.39KK171 pKa = 10.7PPSNAVWPPCKK182 pKa = 9.78RR183 pKa = 11.84SIYY186 pKa = 8.45FHH188 pKa = 7.11KK189 pKa = 10.18FATGLGVKK197 pKa = 7.95TEE199 pKa = 4.21WKK201 pKa = 8.12NTTDD205 pKa = 3.62GGVGSIKK212 pKa = 10.37KK213 pKa = 9.07GALYY217 pKa = 9.47FVIAPGSGIDD227 pKa = 3.5FTLFGTCRR235 pKa = 11.84MYY237 pKa = 10.82FKK239 pKa = 10.94SVGNQQ244 pKa = 2.8
Molecular weight: 26.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
767 |
109 |
266 |
191.8 |
21.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.953 ± 0.271 | 1.956 ± 0.228 |
4.954 ± 0.804 | 5.476 ± 1.221 |
4.433 ± 0.587 | 5.867 ± 2.101 |
2.608 ± 0.671 | 4.433 ± 0.245 |
5.737 ± 0.882 | 7.301 ± 1.525 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.695 ± 0.162 | 3.781 ± 0.831 |
5.997 ± 0.623 | 3.781 ± 0.533 |
5.606 ± 0.643 | 9.648 ± 0.732 |
6.389 ± 0.686 | 6.128 ± 0.858 |
2.347 ± 0.289 | 3.911 ± 0.337 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
