
Mizugakiibacter sediminis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Rhodanobacteraceae; Mizugakiibacter
Average proteome isoelectric point is 7.12
Get precalculated fractions of proteins
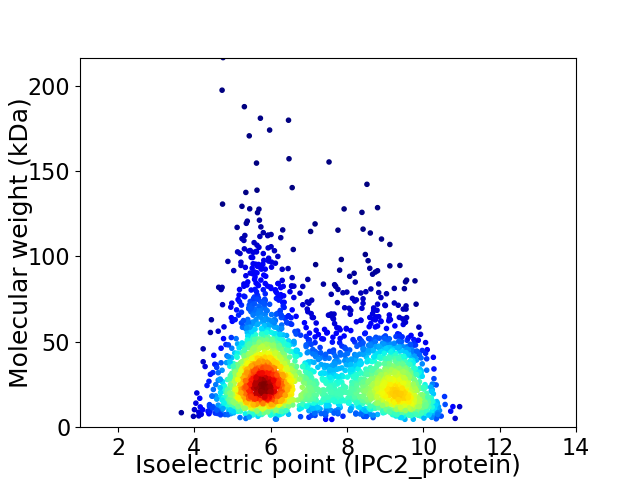
Virtual 2D-PAGE plot for 2837 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
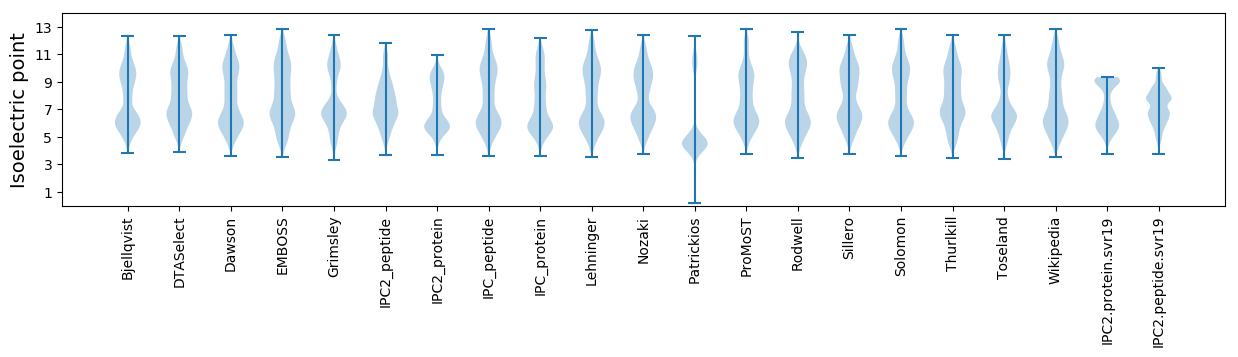
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K8QNG9|A0A0K8QNG9_9GAMM GDP-mannose 4 6-dehydratase OS=Mizugakiibacter sediminis OX=1475481 GN=gmd PE=3 SV=1
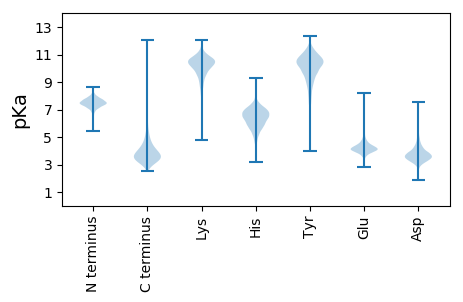
MM1 pKa = 7.16YY2 pKa = 10.08ALSDD6 pKa = 3.24AHH8 pKa = 5.98VQNVSGGGWGDD19 pKa = 4.07AIGAAVVGWAVGKK32 pKa = 10.66VLDD35 pKa = 4.46AAASAIASQNQGGTAFEE52 pKa = 4.96EE53 pKa = 5.08GDD55 pKa = 3.57STLPANVYY63 pKa = 8.06GTT65 pKa = 3.75
MM1 pKa = 7.16YY2 pKa = 10.08ALSDD6 pKa = 3.24AHH8 pKa = 5.98VQNVSGGGWGDD19 pKa = 4.07AIGAAVVGWAVGKK32 pKa = 10.66VLDD35 pKa = 4.46AAASAIASQNQGGTAFEE52 pKa = 4.96EE53 pKa = 5.08GDD55 pKa = 3.57STLPANVYY63 pKa = 8.06GTT65 pKa = 3.75
Molecular weight: 6.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K8QK18|A0A0K8QK18_9GAMM Phe-4-monooxygenase OS=Mizugakiibacter sediminis OX=1475481 GN=MBSD_1711 PE=3 SV=1
MM1 pKa = 7.76AEE3 pKa = 3.85ALADD7 pKa = 3.79GVARR11 pKa = 11.84FLDD14 pKa = 3.59HH15 pKa = 6.87LAVEE19 pKa = 4.97RR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 10.75SPHH25 pKa = 5.91TLAGYY30 pKa = 9.76RR31 pKa = 11.84QDD33 pKa = 3.78LDD35 pKa = 3.71KK36 pKa = 10.97LARR39 pKa = 11.84WMQARR44 pKa = 11.84GLARR48 pKa = 11.84FDD50 pKa = 4.33ALTPDD55 pKa = 4.32LLRR58 pKa = 11.84GFVAAEE64 pKa = 3.86HH65 pKa = 6.49RR66 pKa = 11.84AGLSPKK72 pKa = 9.92SLQRR76 pKa = 11.84VLSTCRR82 pKa = 11.84SLFRR86 pKa = 11.84FLARR90 pKa = 11.84SGALAHH96 pKa = 7.1DD97 pKa = 4.34PAAGVRR103 pKa = 11.84APKK106 pKa = 10.1VRR108 pKa = 11.84RR109 pKa = 11.84KK110 pKa = 10.01LPQVLDD116 pKa = 3.61TDD118 pKa = 3.82EE119 pKa = 5.44ANALVEE125 pKa = 4.41LPAEE129 pKa = 4.52GTLGPRR135 pKa = 11.84DD136 pKa = 3.36RR137 pKa = 11.84AMLEE141 pKa = 4.0LFYY144 pKa = 11.29SSALRR149 pKa = 11.84LSEE152 pKa = 4.68LVGLRR157 pKa = 11.84WGALDD162 pKa = 4.2LDD164 pKa = 4.25EE165 pKa = 5.09GLVRR169 pKa = 11.84VTGKK173 pKa = 10.4GGKK176 pKa = 7.01TRR178 pKa = 11.84IVPVGRR184 pKa = 11.84HH185 pKa = 4.39ACAALRR191 pKa = 11.84ALAAAGGAAPDD202 pKa = 3.56APVFVSRR209 pKa = 11.84HH210 pKa = 4.89GGALTPRR217 pKa = 11.84TVQARR222 pKa = 11.84LKK224 pKa = 9.71TLALRR229 pKa = 11.84QGIAKK234 pKa = 9.97RR235 pKa = 11.84IHH237 pKa = 5.82PHH239 pKa = 6.46LLRR242 pKa = 11.84HH243 pKa = 5.77SCASHH248 pKa = 7.01LLEE251 pKa = 5.06SSGDD255 pKa = 3.51LRR257 pKa = 11.84AVQEE261 pKa = 4.38LLGHH265 pKa = 6.73ADD267 pKa = 2.88IGTTEE272 pKa = 4.59IYY274 pKa = 9.33THH276 pKa = 7.48LDD278 pKa = 3.42FQHH281 pKa = 7.1LARR284 pKa = 11.84VYY286 pKa = 10.55DD287 pKa = 3.9AAHH290 pKa = 5.79PRR292 pKa = 11.84ARR294 pKa = 11.84RR295 pKa = 11.84KK296 pKa = 9.73
MM1 pKa = 7.76AEE3 pKa = 3.85ALADD7 pKa = 3.79GVARR11 pKa = 11.84FLDD14 pKa = 3.59HH15 pKa = 6.87LAVEE19 pKa = 4.97RR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 10.75SPHH25 pKa = 5.91TLAGYY30 pKa = 9.76RR31 pKa = 11.84QDD33 pKa = 3.78LDD35 pKa = 3.71KK36 pKa = 10.97LARR39 pKa = 11.84WMQARR44 pKa = 11.84GLARR48 pKa = 11.84FDD50 pKa = 4.33ALTPDD55 pKa = 4.32LLRR58 pKa = 11.84GFVAAEE64 pKa = 3.86HH65 pKa = 6.49RR66 pKa = 11.84AGLSPKK72 pKa = 9.92SLQRR76 pKa = 11.84VLSTCRR82 pKa = 11.84SLFRR86 pKa = 11.84FLARR90 pKa = 11.84SGALAHH96 pKa = 7.1DD97 pKa = 4.34PAAGVRR103 pKa = 11.84APKK106 pKa = 10.1VRR108 pKa = 11.84RR109 pKa = 11.84KK110 pKa = 10.01LPQVLDD116 pKa = 3.61TDD118 pKa = 3.82EE119 pKa = 5.44ANALVEE125 pKa = 4.41LPAEE129 pKa = 4.52GTLGPRR135 pKa = 11.84DD136 pKa = 3.36RR137 pKa = 11.84AMLEE141 pKa = 4.0LFYY144 pKa = 11.29SSALRR149 pKa = 11.84LSEE152 pKa = 4.68LVGLRR157 pKa = 11.84WGALDD162 pKa = 4.2LDD164 pKa = 4.25EE165 pKa = 5.09GLVRR169 pKa = 11.84VTGKK173 pKa = 10.4GGKK176 pKa = 7.01TRR178 pKa = 11.84IVPVGRR184 pKa = 11.84HH185 pKa = 4.39ACAALRR191 pKa = 11.84ALAAAGGAAPDD202 pKa = 3.56APVFVSRR209 pKa = 11.84HH210 pKa = 4.89GGALTPRR217 pKa = 11.84TVQARR222 pKa = 11.84LKK224 pKa = 9.71TLALRR229 pKa = 11.84QGIAKK234 pKa = 9.97RR235 pKa = 11.84IHH237 pKa = 5.82PHH239 pKa = 6.46LLRR242 pKa = 11.84HH243 pKa = 5.77SCASHH248 pKa = 7.01LLEE251 pKa = 5.06SSGDD255 pKa = 3.51LRR257 pKa = 11.84AVQEE261 pKa = 4.38LLGHH265 pKa = 6.73ADD267 pKa = 2.88IGTTEE272 pKa = 4.59IYY274 pKa = 9.33THH276 pKa = 7.48LDD278 pKa = 3.42FQHH281 pKa = 7.1LARR284 pKa = 11.84VYY286 pKa = 10.55DD287 pKa = 3.9AAHH290 pKa = 5.79PRR292 pKa = 11.84ARR294 pKa = 11.84RR295 pKa = 11.84KK296 pKa = 9.73
Molecular weight: 32.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
897279 |
39 |
1999 |
316.3 |
34.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.007 ± 0.096 | 0.901 ± 0.015 |
5.65 ± 0.037 | 5.424 ± 0.047 |
3.318 ± 0.027 | 8.526 ± 0.036 |
2.284 ± 0.023 | 3.779 ± 0.033 |
2.569 ± 0.04 | 11.18 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.009 ± 0.02 | 2.044 ± 0.028 |
5.553 ± 0.033 | 3.211 ± 0.027 |
8.653 ± 0.054 | 4.118 ± 0.033 |
4.52 ± 0.035 | 7.374 ± 0.037 |
1.48 ± 0.021 | 2.401 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
