
Sediminibacterium goheungense
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Sediminibacterium
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
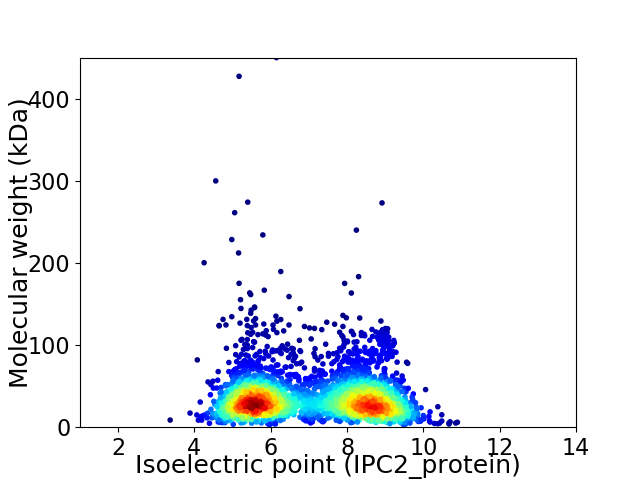
Virtual 2D-PAGE plot for 3224 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6ITU4|A0A4R6ITU4_9BACT Sortilin (Neurotensin receptor 3) OS=Sediminibacterium goheungense OX=1086393 GN=BC659_2650 PE=4 SV=1
MM1 pKa = 7.6NKK3 pKa = 8.21FTRR6 pKa = 11.84LWVSLVSLTLFTVFSAFRR24 pKa = 11.84FTGLPEE30 pKa = 4.3LGGAEE35 pKa = 4.08KK36 pKa = 10.03MYY38 pKa = 10.72NNPGKK43 pKa = 9.94VVKK46 pKa = 10.43DD47 pKa = 3.25ATVVRR52 pKa = 11.84NFTTRR57 pKa = 11.84VLKK60 pKa = 10.9NITRR64 pKa = 11.84AAGFTGRR71 pKa = 11.84TFNGNSNISLTVPGNMSFSNTPGTCGRR98 pKa = 11.84VVTYY102 pKa = 9.74AAPTAQANAVSVEE115 pKa = 3.63FDD117 pKa = 3.31YY118 pKa = 11.3TGNIQTFTVPAGVTSITIKK137 pKa = 11.03GIGADD142 pKa = 3.79GGNSLSSGVNGGRR155 pKa = 11.84GASLTGTFAVTPGQTIYY172 pKa = 10.46MVVGQRR178 pKa = 11.84GSDD181 pKa = 3.5DD182 pKa = 3.65FADD185 pKa = 4.71FGLGAAGGGGGTYY198 pKa = 10.06ISATPFGTPSAVPLMVAGAGGGAAAIVDD226 pKa = 4.07NNIHH230 pKa = 6.57ANANSVDD237 pKa = 3.5GNYY240 pKa = 10.73GVTGDD245 pKa = 4.55GGDD248 pKa = 3.66GALGTGGSGGEE259 pKa = 3.93AGTNSGAGAGWLADD273 pKa = 3.64GLSDD277 pKa = 4.92INSGDD282 pKa = 3.45LTGGLSQFGTNPFAGGSLFGVLYY305 pKa = 10.53GGYY308 pKa = 10.23GGGGSAGFVGGGGGGGYY325 pKa = 9.95SGGGGGGDD333 pKa = 3.14NGGGGGGGSFNGGTNPVNVPGVDD356 pKa = 3.59GDD358 pKa = 4.63DD359 pKa = 3.71GAGGNGKK366 pKa = 9.04VVITYY371 pKa = 7.42GTSVTVTQIAGLPSGATFPVGVTTNTFQATDD402 pKa = 3.74GVNTTTASFTVTVNDD417 pKa = 3.52TEE419 pKa = 5.0APVLTAPSNITYY431 pKa = 7.79TIPAGSSNCNAAAVNLGTPTVSDD454 pKa = 3.27NCGANTPTNNAPSLFPLGTTTVTWSVTDD482 pKa = 2.82IHH484 pKa = 6.99GNPATATQQVTVVAPEE500 pKa = 3.64IRR502 pKa = 11.84IASASFVFSILNGSTSIDD520 pKa = 3.73LATGTDD526 pKa = 3.98FGTTNINNQVTRR538 pKa = 11.84TYY540 pKa = 10.46IIDD543 pKa = 3.81NAGGTADD550 pKa = 4.83LSVNSINIAGANPSFFTVGGITLPATIPAGQTTTFTVTYY589 pKa = 8.94SASAGGTHH597 pKa = 6.44NAIVQLNNNDD607 pKa = 3.93CDD609 pKa = 3.39EE610 pKa = 4.39SAYY613 pKa = 11.1SFAIQGVTNQATTPITGNTSVCVSGTTQLANVTAGGTWSSANAAVATVDD662 pKa = 3.68ANGLVTAVSAGTANISYY679 pKa = 8.18TMPGGTVQVTVTVNAAPTAAISANGPTTFCAGGSVTLTATGGASYY724 pKa = 10.35LWSNGSTTSSITVSAAGTYY743 pKa = 10.41SVVATSAEE751 pKa = 4.4GCSSASVSSSVTVNPVPTAAISADD775 pKa = 3.52GPTTFCAGGSVTLTATGGASYY796 pKa = 10.28LWSNGATTSSITVSSGGVYY815 pKa = 10.34SVVATSAEE823 pKa = 4.4GCSSASVSSSVTVNPVPTAAISAA846 pKa = 4.02
MM1 pKa = 7.6NKK3 pKa = 8.21FTRR6 pKa = 11.84LWVSLVSLTLFTVFSAFRR24 pKa = 11.84FTGLPEE30 pKa = 4.3LGGAEE35 pKa = 4.08KK36 pKa = 10.03MYY38 pKa = 10.72NNPGKK43 pKa = 9.94VVKK46 pKa = 10.43DD47 pKa = 3.25ATVVRR52 pKa = 11.84NFTTRR57 pKa = 11.84VLKK60 pKa = 10.9NITRR64 pKa = 11.84AAGFTGRR71 pKa = 11.84TFNGNSNISLTVPGNMSFSNTPGTCGRR98 pKa = 11.84VVTYY102 pKa = 9.74AAPTAQANAVSVEE115 pKa = 3.63FDD117 pKa = 3.31YY118 pKa = 11.3TGNIQTFTVPAGVTSITIKK137 pKa = 11.03GIGADD142 pKa = 3.79GGNSLSSGVNGGRR155 pKa = 11.84GASLTGTFAVTPGQTIYY172 pKa = 10.46MVVGQRR178 pKa = 11.84GSDD181 pKa = 3.5DD182 pKa = 3.65FADD185 pKa = 4.71FGLGAAGGGGGTYY198 pKa = 10.06ISATPFGTPSAVPLMVAGAGGGAAAIVDD226 pKa = 4.07NNIHH230 pKa = 6.57ANANSVDD237 pKa = 3.5GNYY240 pKa = 10.73GVTGDD245 pKa = 4.55GGDD248 pKa = 3.66GALGTGGSGGEE259 pKa = 3.93AGTNSGAGAGWLADD273 pKa = 3.64GLSDD277 pKa = 4.92INSGDD282 pKa = 3.45LTGGLSQFGTNPFAGGSLFGVLYY305 pKa = 10.53GGYY308 pKa = 10.23GGGGSAGFVGGGGGGGYY325 pKa = 9.95SGGGGGGDD333 pKa = 3.14NGGGGGGGSFNGGTNPVNVPGVDD356 pKa = 3.59GDD358 pKa = 4.63DD359 pKa = 3.71GAGGNGKK366 pKa = 9.04VVITYY371 pKa = 7.42GTSVTVTQIAGLPSGATFPVGVTTNTFQATDD402 pKa = 3.74GVNTTTASFTVTVNDD417 pKa = 3.52TEE419 pKa = 5.0APVLTAPSNITYY431 pKa = 7.79TIPAGSSNCNAAAVNLGTPTVSDD454 pKa = 3.27NCGANTPTNNAPSLFPLGTTTVTWSVTDD482 pKa = 2.82IHH484 pKa = 6.99GNPATATQQVTVVAPEE500 pKa = 3.64IRR502 pKa = 11.84IASASFVFSILNGSTSIDD520 pKa = 3.73LATGTDD526 pKa = 3.98FGTTNINNQVTRR538 pKa = 11.84TYY540 pKa = 10.46IIDD543 pKa = 3.81NAGGTADD550 pKa = 4.83LSVNSINIAGANPSFFTVGGITLPATIPAGQTTTFTVTYY589 pKa = 8.94SASAGGTHH597 pKa = 6.44NAIVQLNNNDD607 pKa = 3.93CDD609 pKa = 3.39EE610 pKa = 4.39SAYY613 pKa = 11.1SFAIQGVTNQATTPITGNTSVCVSGTTQLANVTAGGTWSSANAAVATVDD662 pKa = 3.68ANGLVTAVSAGTANISYY679 pKa = 8.18TMPGGTVQVTVTVNAAPTAAISANGPTTFCAGGSVTLTATGGASYY724 pKa = 10.35LWSNGSTTSSITVSAAGTYY743 pKa = 10.41SVVATSAEE751 pKa = 4.4GCSSASVSSSVTVNPVPTAAISADD775 pKa = 3.52GPTTFCAGGSVTLTATGGASYY796 pKa = 10.28LWSNGATTSSITVSSGGVYY815 pKa = 10.34SVVATSAEE823 pKa = 4.4GCSSASVSSSVTVNPVPTAAISAA846 pKa = 4.02
Molecular weight: 81.91 kDa
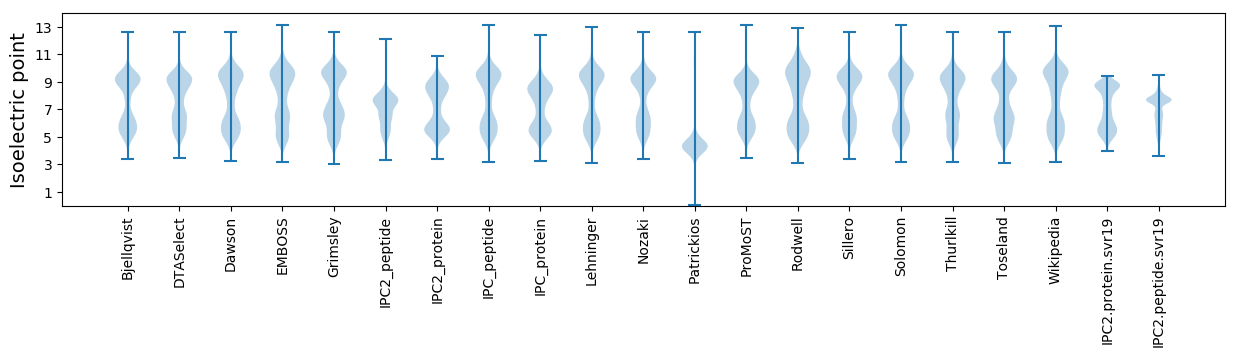
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6IWM2|A0A4R6IWM2_9BACT NIPSNAP protein OS=Sediminibacterium goheungense OX=1086393 GN=BC659_2423 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 9.59RR3 pKa = 11.84TFQPHH8 pKa = 4.67KK9 pKa = 9.7RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.4SVHH16 pKa = 5.0GFRR19 pKa = 11.84KK20 pKa = 10.11RR21 pKa = 11.84MQTANGRR28 pKa = 11.84KK29 pKa = 8.71VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.04GRR39 pKa = 11.84KK40 pKa = 8.7KK41 pKa = 10.08LTVSSEE47 pKa = 4.1KK48 pKa = 10.7GLKK51 pKa = 9.34
MM1 pKa = 7.38KK2 pKa = 9.59RR3 pKa = 11.84TFQPHH8 pKa = 4.67KK9 pKa = 9.7RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.4SVHH16 pKa = 5.0GFRR19 pKa = 11.84KK20 pKa = 10.11RR21 pKa = 11.84MQTANGRR28 pKa = 11.84KK29 pKa = 8.71VLASRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.04GRR39 pKa = 11.84KK40 pKa = 8.7KK41 pKa = 10.08LTVSSEE47 pKa = 4.1KK48 pKa = 10.7GLKK51 pKa = 9.34
Molecular weight: 6.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1116394 |
29 |
4357 |
346.3 |
38.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.563 ± 0.044 | 0.878 ± 0.014 |
5.046 ± 0.037 | 5.617 ± 0.062 |
4.881 ± 0.033 | 6.877 ± 0.042 |
1.94 ± 0.025 | 7.288 ± 0.04 |
6.85 ± 0.052 | 9.342 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.503 ± 0.026 | 5.312 ± 0.054 |
3.85 ± 0.025 | 4.034 ± 0.029 |
4.112 ± 0.03 | 6.261 ± 0.05 |
6.024 ± 0.081 | 6.36 ± 0.032 |
1.272 ± 0.017 | 3.992 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
