
Alienimonas californiensis
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; Alienimonas
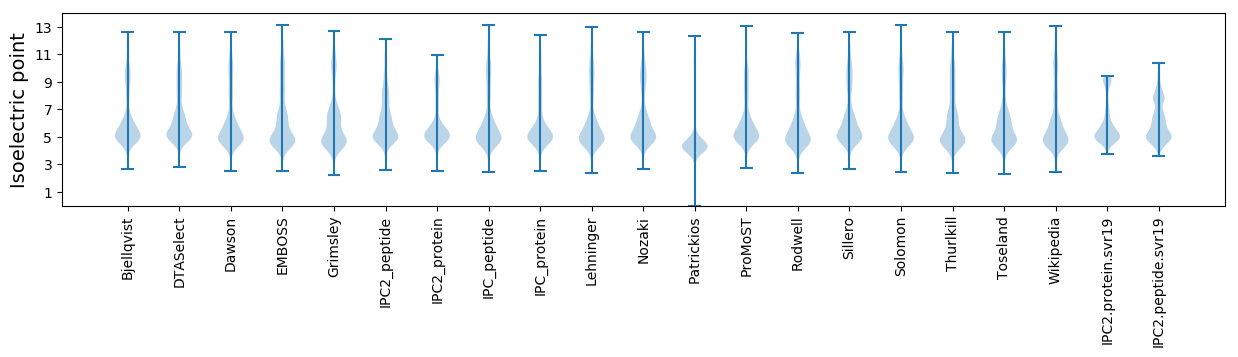
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
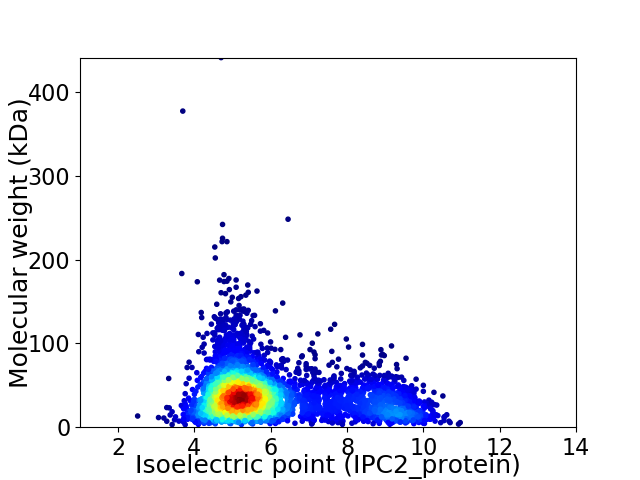
Virtual 2D-PAGE plot for 4303 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A517P485|A0A517P485_9PLAN Uncharacterized protein OS=Alienimonas californiensis OX=2527989 GN=CA12_02660 PE=4 SV=1
MM1 pKa = 7.57LPPLKK6 pKa = 10.34RR7 pKa = 11.84HH8 pKa = 6.06ADD10 pKa = 3.71RR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84AAVLGIVAGGAATPAFAGPPPCPPPAYY41 pKa = 9.9NPCPPAYY48 pKa = 9.64NPCPPGVITPVTPAPPAPMDD68 pKa = 4.08GSATEE73 pKa = 4.39LLPEE77 pKa = 4.42TFSRR81 pKa = 11.84TAPPSPAAQNLVANTANPLAGRR103 pKa = 11.84SGLAAVGGGVGDD115 pKa = 4.28PGYY118 pKa = 10.34IDD120 pKa = 3.82PAVVASRR127 pKa = 11.84FRR129 pKa = 11.84LRR131 pKa = 11.84YY132 pKa = 9.7SGARR136 pKa = 11.84DD137 pKa = 3.49GNSDD141 pKa = 3.43RR142 pKa = 11.84GEE144 pKa = 4.25FLYY147 pKa = 10.74GAYY150 pKa = 7.94DD151 pKa = 3.86TPALQASLPPGTPAVFIGPGNPGNVAAVPGQTVSGPTGPVRR192 pKa = 11.84PVIDD196 pKa = 3.84GFDD199 pKa = 3.54YY200 pKa = 11.54DD201 pKa = 4.66EE202 pKa = 4.51ISAYY206 pKa = 10.85LEE208 pKa = 4.18LEE210 pKa = 4.19VVEE213 pKa = 4.66GVSAFVEE220 pKa = 4.89VPWRR224 pKa = 11.84DD225 pKa = 3.17VSGYY229 pKa = 10.5DD230 pKa = 3.17QFRR233 pKa = 11.84DD234 pKa = 3.41QGFADD239 pKa = 5.35DD240 pKa = 5.32GLGDD244 pKa = 3.39INAGVRR250 pKa = 11.84VSLYY254 pKa = 10.62DD255 pKa = 3.88DD256 pKa = 4.97CDD258 pKa = 3.5TALTFQLMYY267 pKa = 9.04QTNTGDD273 pKa = 3.61PDD275 pKa = 3.88FGLGNGTQFIVPGLLFQQSLSDD297 pKa = 3.9DD298 pKa = 3.31LHH300 pKa = 8.08LFGEE304 pKa = 4.22AQYY307 pKa = 11.39YY308 pKa = 9.67VDD310 pKa = 4.81AGNGTEE316 pKa = 5.47FNGEE320 pKa = 4.4DD321 pKa = 3.59YY322 pKa = 11.0TSDD325 pKa = 3.18VLRR328 pKa = 11.84FGLGAALDD336 pKa = 4.02LVEE339 pKa = 4.79TRR341 pKa = 11.84DD342 pKa = 3.99TNLQLLNEE350 pKa = 4.31FVGWTFYY357 pKa = 11.07DD358 pKa = 4.09GQQSEE363 pKa = 4.95FNPATRR369 pKa = 11.84QIDD372 pKa = 3.66VTDD375 pKa = 4.21ADD377 pKa = 4.21GEE379 pKa = 4.71TIVNYY384 pKa = 10.24KK385 pKa = 10.27VGLRR389 pKa = 11.84YY390 pKa = 10.02LFGRR394 pKa = 11.84QSVYY398 pKa = 10.98AGYY401 pKa = 11.05GLALTDD407 pKa = 4.25DD408 pKa = 4.02VLYY411 pKa = 11.01DD412 pKa = 4.21DD413 pKa = 4.8IVRR416 pKa = 11.84LEE418 pKa = 3.89YY419 pKa = 10.75SVLLDD424 pKa = 3.45
MM1 pKa = 7.57LPPLKK6 pKa = 10.34RR7 pKa = 11.84HH8 pKa = 6.06ADD10 pKa = 3.71RR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84AAVLGIVAGGAATPAFAGPPPCPPPAYY41 pKa = 9.9NPCPPAYY48 pKa = 9.64NPCPPGVITPVTPAPPAPMDD68 pKa = 4.08GSATEE73 pKa = 4.39LLPEE77 pKa = 4.42TFSRR81 pKa = 11.84TAPPSPAAQNLVANTANPLAGRR103 pKa = 11.84SGLAAVGGGVGDD115 pKa = 4.28PGYY118 pKa = 10.34IDD120 pKa = 3.82PAVVASRR127 pKa = 11.84FRR129 pKa = 11.84LRR131 pKa = 11.84YY132 pKa = 9.7SGARR136 pKa = 11.84DD137 pKa = 3.49GNSDD141 pKa = 3.43RR142 pKa = 11.84GEE144 pKa = 4.25FLYY147 pKa = 10.74GAYY150 pKa = 7.94DD151 pKa = 3.86TPALQASLPPGTPAVFIGPGNPGNVAAVPGQTVSGPTGPVRR192 pKa = 11.84PVIDD196 pKa = 3.84GFDD199 pKa = 3.54YY200 pKa = 11.54DD201 pKa = 4.66EE202 pKa = 4.51ISAYY206 pKa = 10.85LEE208 pKa = 4.18LEE210 pKa = 4.19VVEE213 pKa = 4.66GVSAFVEE220 pKa = 4.89VPWRR224 pKa = 11.84DD225 pKa = 3.17VSGYY229 pKa = 10.5DD230 pKa = 3.17QFRR233 pKa = 11.84DD234 pKa = 3.41QGFADD239 pKa = 5.35DD240 pKa = 5.32GLGDD244 pKa = 3.39INAGVRR250 pKa = 11.84VSLYY254 pKa = 10.62DD255 pKa = 3.88DD256 pKa = 4.97CDD258 pKa = 3.5TALTFQLMYY267 pKa = 9.04QTNTGDD273 pKa = 3.61PDD275 pKa = 3.88FGLGNGTQFIVPGLLFQQSLSDD297 pKa = 3.9DD298 pKa = 3.31LHH300 pKa = 8.08LFGEE304 pKa = 4.22AQYY307 pKa = 11.39YY308 pKa = 9.67VDD310 pKa = 4.81AGNGTEE316 pKa = 5.47FNGEE320 pKa = 4.4DD321 pKa = 3.59YY322 pKa = 11.0TSDD325 pKa = 3.18VLRR328 pKa = 11.84FGLGAALDD336 pKa = 4.02LVEE339 pKa = 4.79TRR341 pKa = 11.84DD342 pKa = 3.99TNLQLLNEE350 pKa = 4.31FVGWTFYY357 pKa = 11.07DD358 pKa = 4.09GQQSEE363 pKa = 4.95FNPATRR369 pKa = 11.84QIDD372 pKa = 3.66VTDD375 pKa = 4.21ADD377 pKa = 4.21GEE379 pKa = 4.71TIVNYY384 pKa = 10.24KK385 pKa = 10.27VGLRR389 pKa = 11.84YY390 pKa = 10.02LFGRR394 pKa = 11.84QSVYY398 pKa = 10.98AGYY401 pKa = 11.05GLALTDD407 pKa = 4.25DD408 pKa = 4.02VLYY411 pKa = 11.01DD412 pKa = 4.21DD413 pKa = 4.8IVRR416 pKa = 11.84LEE418 pKa = 3.89YY419 pKa = 10.75SVLLDD424 pKa = 3.45
Molecular weight: 44.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A517P9L4|A0A517P9L4_9PLAN Putative major pilin subunit OS=Alienimonas californiensis OX=2527989 GN=CA12_21610 PE=4 SV=1
MM1 pKa = 7.83AKK3 pKa = 8.26TQRR6 pKa = 11.84KK7 pKa = 8.43LKK9 pKa = 9.62KK10 pKa = 8.39AHH12 pKa = 6.49HH13 pKa = 6.56GSRR16 pKa = 11.84PASSKK21 pKa = 10.28ARR23 pKa = 11.84KK24 pKa = 8.05QKK26 pKa = 10.21RR27 pKa = 11.84RR28 pKa = 11.84AIKK31 pKa = 8.92TT32 pKa = 3.46
MM1 pKa = 7.83AKK3 pKa = 8.26TQRR6 pKa = 11.84KK7 pKa = 8.43LKK9 pKa = 9.62KK10 pKa = 8.39AHH12 pKa = 6.49HH13 pKa = 6.56GSRR16 pKa = 11.84PASSKK21 pKa = 10.28ARR23 pKa = 11.84KK24 pKa = 8.05QKK26 pKa = 10.21RR27 pKa = 11.84RR28 pKa = 11.84AIKK31 pKa = 8.92TT32 pKa = 3.46
Molecular weight: 3.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1597559 |
30 |
4167 |
371.3 |
39.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.874 ± 0.068 | 1.001 ± 0.015 |
6.343 ± 0.026 | 6.29 ± 0.035 |
3.323 ± 0.022 | 9.349 ± 0.04 |
1.899 ± 0.019 | 2.949 ± 0.023 |
2.479 ± 0.031 | 10.165 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.556 ± 0.017 | 2.221 ± 0.021 |
6.925 ± 0.041 | 2.491 ± 0.019 |
7.911 ± 0.041 | 4.801 ± 0.023 |
5.394 ± 0.026 | 7.58 ± 0.033 |
1.486 ± 0.016 | 1.963 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
