
Pedobacter sp. Leaf216
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Sphingobacteriia; Sphingobacteriales; Sphingobacteriaceae; Pedobacter; unclassified Pedobacter
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
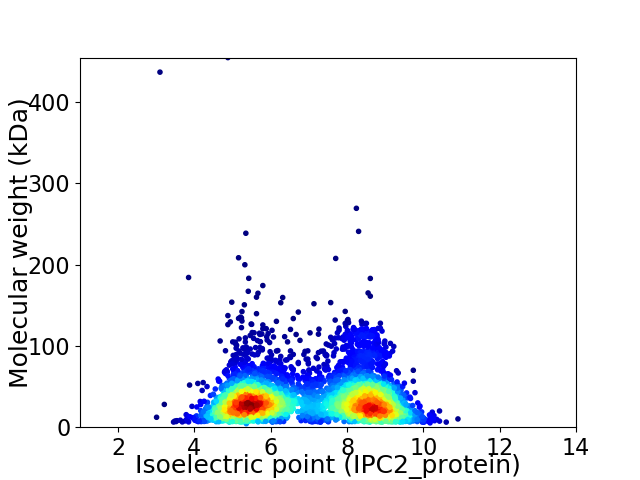
Virtual 2D-PAGE plot for 4730 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q4G7J9|A0A0Q4G7J9_9SPHI Sodium:solute symporter OS=Pedobacter sp. Leaf216 OX=1735684 GN=ASE74_05085 PE=3 SV=1
MM1 pKa = 7.67INPDD5 pKa = 2.86GTYY8 pKa = 9.94TYY10 pKa = 10.56TPAPNYY16 pKa = 10.61NGTDD20 pKa = 3.31SFTVTVSDD28 pKa = 3.99GKK30 pKa = 11.28GGTTTVTINVTITPVNDD47 pKa = 3.48APVATAPAIVTNKK60 pKa = 9.91NIPVNGTITASDD72 pKa = 3.54VDD74 pKa = 4.0GDD76 pKa = 4.09ALTFTVSTPPAHH88 pKa = 5.69GTVVVKK94 pKa = 10.88ADD96 pKa = 3.27GTYY99 pKa = 10.2TYY101 pKa = 10.32TPANNYY107 pKa = 9.78SGSDD111 pKa = 3.45VFTVTVSDD119 pKa = 4.07GKK121 pKa = 11.22GGTATVTINVTINPTNVAPVVTAPAVTGNEE151 pKa = 3.95DD152 pKa = 3.45SPINGTITATDD163 pKa = 3.62ADD165 pKa = 4.47GDD167 pKa = 4.2PLTFTVATPPSNGTVVVNPDD187 pKa = 2.68GTYY190 pKa = 10.12TYY192 pKa = 10.56TPAPNYY198 pKa = 10.61NGTDD202 pKa = 3.31SFTVTVSDD210 pKa = 3.99GKK212 pKa = 11.28GGTTTVTVNVNVTPVNDD229 pKa = 3.77APVATSPAIVTNKK242 pKa = 9.94NIPVNGTITASDD254 pKa = 3.54VDD256 pKa = 4.0GDD258 pKa = 4.09ALTFTVSTPPAHH270 pKa = 5.76GTVVVNADD278 pKa = 3.0GTYY281 pKa = 10.14TYY283 pKa = 10.48TPANNYY289 pKa = 9.78SGSDD293 pKa = 3.45VFTVTVSDD301 pKa = 3.96GKK303 pKa = 11.28GGTTTVTINVTVNPTNVAPVVTAPAVTGNEE333 pKa = 3.95DD334 pKa = 3.45SPINGTITATDD345 pKa = 3.62ADD347 pKa = 4.47GDD349 pKa = 4.2PLTFTVATPPANGTVVVNPDD369 pKa = 2.68GTYY372 pKa = 10.15TYY374 pKa = 10.66TPVPNYY380 pKa = 10.71NGTDD384 pKa = 3.37SFTVTVSDD392 pKa = 3.99GKK394 pKa = 11.28GGTTTVTINVTVTPVNDD411 pKa = 3.82APVATAPAIVTNKK424 pKa = 9.91NIPVNGTITASDD436 pKa = 3.54VDD438 pKa = 4.0GDD440 pKa = 4.09ALTFTVSTPPAHH452 pKa = 5.76GTVVVNADD460 pKa = 3.0GTYY463 pKa = 10.14TYY465 pKa = 10.48TPANNYY471 pKa = 9.78SGSDD475 pKa = 3.45VFTVTVSDD483 pKa = 3.96GKK485 pKa = 11.28GGTTTVTINVTINPTNVAPVVTAPAVTGNEE515 pKa = 3.95DD516 pKa = 3.45SPINGTITATDD527 pKa = 3.62ADD529 pKa = 4.47GDD531 pKa = 4.22PLTFTVSTPPAHH543 pKa = 5.66GTVVINPDD551 pKa = 2.67GTYY554 pKa = 10.01TYY556 pKa = 10.56TPAPNYY562 pKa = 10.61NGTDD566 pKa = 3.31SFTVTVSDD574 pKa = 3.99GKK576 pKa = 11.28GGTTTVTINVTVTPPEE592 pKa = 4.59TISXTPPAHH601 pKa = 5.29GTVVINPDD609 pKa = 2.67GTYY612 pKa = 10.01TYY614 pKa = 10.56TPAPNYY620 pKa = 10.61NGTDD624 pKa = 3.31SFTVTVSDD632 pKa = 3.99GKK634 pKa = 11.28GGTTTVTINVIINPVNDD651 pKa = 3.98APVATAPAIVTNKK664 pKa = 9.91NIPVNGTITASDD676 pKa = 3.54VDD678 pKa = 4.0GDD680 pKa = 4.09ALTFTVSTPPAHH692 pKa = 6.07GTVTVNADD700 pKa = 2.98GTYY703 pKa = 10.12TYY705 pKa = 10.48TPANNYY711 pKa = 9.78SGSDD715 pKa = 3.45VFTVTVSDD723 pKa = 4.07GKK725 pKa = 11.22GGTATVTINVTINPTNVAPVVTAPAVTGNEE755 pKa = 3.95DD756 pKa = 3.45SPINGTITATDD767 pKa = 3.62ADD769 pKa = 4.47GDD771 pKa = 4.2PLTFTVATPPAHH783 pKa = 5.67GTVTVNADD791 pKa = 2.98GTYY794 pKa = 9.93TYY796 pKa = 10.65TPAPNYY802 pKa = 10.61NGTDD806 pKa = 3.31SFTVTVSDD814 pKa = 3.99GKK816 pKa = 11.28GGTTTVTVNVNVTPVNDD833 pKa = 3.77APVATSPAIVTNKK846 pKa = 9.94NIPVNGTITASDD858 pKa = 3.54VDD860 pKa = 4.0GDD862 pKa = 4.09ALTFTVSTPPAHH874 pKa = 6.07GTVTVNADD882 pKa = 2.98GTYY885 pKa = 10.12TYY887 pKa = 10.48TPANNYY893 pKa = 9.78SGSDD897 pKa = 3.45VFTVTVSDD905 pKa = 3.96GKK907 pKa = 11.28GGTTTVTINVTVNPTNVAPFVTSPAVTGNEE937 pKa = 3.88DD938 pKa = 3.46SPINSSITATDD949 pKa = 3.53ADD951 pKa = 4.4GDD953 pKa = 4.22PLTFTVSTPPAHH965 pKa = 5.83GTVVVNPDD973 pKa = 2.68GTYY976 pKa = 10.12TYY978 pKa = 10.56TPAPNYY984 pKa = 10.61NGTDD988 pKa = 3.31SFTVTVSDD996 pKa = 3.99GKK998 pKa = 11.28GGTTTVTVNVTVTPVNDD1015 pKa = 3.61APVATSPAIVTNKK1028 pKa = 9.94NIPVNGTITASDD1040 pKa = 3.54VDD1042 pKa = 4.0GDD1044 pKa = 4.09ALTFTVSTPPAHH1056 pKa = 5.91GTVTVKK1062 pKa = 10.95ADD1064 pKa = 3.24GTYY1067 pKa = 10.18TYY1069 pKa = 10.32TPANNYY1075 pKa = 9.78SGSDD1079 pKa = 3.45VFTVTVSDD1087 pKa = 3.96GKK1089 pKa = 11.28GGTTTVTINVTINPTNVAPVVTAPAVTGNEE1119 pKa = 3.95DD1120 pKa = 3.45SPINGTITATDD1131 pKa = 3.6ADD1133 pKa = 4.51GDD1135 pKa = 4.12PLIFTVATPPAHH1147 pKa = 5.67GTVTVNADD1155 pKa = 3.02GTYY1158 pKa = 10.07TYY1160 pKa = 10.69IPAPNYY1166 pKa = 10.61NGTDD1170 pKa = 3.31SFTVTVSDD1178 pKa = 3.99GKK1180 pKa = 11.28GGTTTVTVNVTVTPVNDD1197 pKa = 3.91APVTTAPAVTTPQNTAVNGAITASDD1222 pKa = 3.54VDD1224 pKa = 3.93GDD1226 pKa = 4.21ALTFTLTAQPAHH1238 pKa = 5.74GTVVVNANGTYY1249 pKa = 9.83TYY1251 pKa = 9.89TPTAGYY1257 pKa = 9.76SGNDD1261 pKa = 3.09TFTITVSDD1269 pKa = 3.94GKK1271 pKa = 10.97GGTVTVTVPVTVTLVAAPAMSLTKK1295 pKa = 9.92TATNTVSKK1303 pKa = 10.81VGDD1306 pKa = 3.61VINYY1310 pKa = 9.36NIIVTNTGNVTLTNVAVTDD1329 pKa = 4.21AGADD1333 pKa = 3.51AGSLTPAGIASLLPGSSVTVTARR1356 pKa = 11.84HH1357 pKa = 5.22TVTLTEE1363 pKa = 4.14VNAGSFSNQASATAQTPGGSILNKK1387 pKa = 9.85PKK1389 pKa = 10.6SDD1391 pKa = 4.18DD1392 pKa = 3.85PNTIAVDD1399 pKa = 3.75DD1400 pKa = 4.06ATVTVIAPASMITLVKK1416 pKa = 9.58TGTVSTDD1423 pKa = 3.12GNSITYY1429 pKa = 9.97SFSVKK1434 pKa = 8.97NTGNVTLHH1442 pKa = 7.06IITLVDD1448 pKa = 3.54AKK1450 pKa = 10.93LGLNRR1455 pKa = 11.84VIPGTLAPGATVTDD1469 pKa = 3.86SYY1471 pKa = 11.75VYY1473 pKa = 10.74QLTQADD1479 pKa = 3.97KK1480 pKa = 11.31DD1481 pKa = 3.84AGSVTNTASITGQTPANVAVSDD1503 pKa = 3.65ISGTAEE1509 pKa = 3.86NNNTPTVTLIPNNGSVALVKK1529 pKa = 10.16TSVFNGNKK1537 pKa = 6.32VTYY1540 pKa = 9.59TFTIKK1545 pKa = 9.56NTGTITLNTITLTDD1559 pKa = 3.69AKK1561 pKa = 10.69LGLNNKK1567 pKa = 9.2VITVAGGLAPGATTTDD1583 pKa = 3.41VEE1585 pKa = 5.23VYY1587 pKa = 10.12TLTQADD1593 pKa = 4.13KK1594 pKa = 11.53DD1595 pKa = 4.02LGTVTNTATVNARR1608 pKa = 11.84TLAGASVTDD1617 pKa = 3.32VSGTAEE1623 pKa = 4.27TNNTPTVTTFPKK1635 pKa = 10.65SPTAVDD1641 pKa = 4.71DD1642 pKa = 4.12KK1643 pKa = 11.36AQTVANSPVVISVLNNDD1660 pKa = 4.15DD1661 pKa = 4.74PGNSTLDD1668 pKa = 3.41KK1669 pKa = 10.54LTVEE1673 pKa = 4.55VVNAPKK1679 pKa = 10.54HH1680 pKa = 4.79GTVKK1684 pKa = 10.49VNADD1688 pKa = 3.28GTVTYY1693 pKa = 9.57TPDD1696 pKa = 3.5PGYY1699 pKa = 11.22VGDD1702 pKa = 5.2DD1703 pKa = 3.23VFTYY1707 pKa = 10.06RR1708 pKa = 11.84VKK1710 pKa = 10.74DD1711 pKa = 3.42AYY1713 pKa = 10.55GYY1715 pKa = 6.52YY1716 pKa = 10.08TNVATVTLTANFTGITVPNLFTPNGDD1742 pKa = 3.95GINDD1746 pKa = 3.63TFEE1749 pKa = 5.85IIGLNQYY1756 pKa = 8.03QVNEE1760 pKa = 3.97LQIVNRR1766 pKa = 11.84WGNEE1770 pKa = 3.45VFHH1773 pKa = 7.28AKK1775 pKa = 10.2GYY1777 pKa = 8.02QNNWTGEE1784 pKa = 3.99GLNEE1788 pKa = 3.67GTYY1791 pKa = 10.78YY1792 pKa = 11.08YY1793 pKa = 11.2LLRR1796 pKa = 11.84VKK1798 pKa = 10.45KK1799 pKa = 10.38AGSDD1803 pKa = 3.25QYY1805 pKa = 11.61EE1806 pKa = 4.23VFKK1809 pKa = 11.09GYY1811 pKa = 8.32ITLIKK1816 pKa = 10.31AFKK1819 pKa = 10.15KK1820 pKa = 10.59
MM1 pKa = 7.67INPDD5 pKa = 2.86GTYY8 pKa = 9.94TYY10 pKa = 10.56TPAPNYY16 pKa = 10.61NGTDD20 pKa = 3.31SFTVTVSDD28 pKa = 3.99GKK30 pKa = 11.28GGTTTVTINVTITPVNDD47 pKa = 3.48APVATAPAIVTNKK60 pKa = 9.91NIPVNGTITASDD72 pKa = 3.54VDD74 pKa = 4.0GDD76 pKa = 4.09ALTFTVSTPPAHH88 pKa = 5.69GTVVVKK94 pKa = 10.88ADD96 pKa = 3.27GTYY99 pKa = 10.2TYY101 pKa = 10.32TPANNYY107 pKa = 9.78SGSDD111 pKa = 3.45VFTVTVSDD119 pKa = 4.07GKK121 pKa = 11.22GGTATVTINVTINPTNVAPVVTAPAVTGNEE151 pKa = 3.95DD152 pKa = 3.45SPINGTITATDD163 pKa = 3.62ADD165 pKa = 4.47GDD167 pKa = 4.2PLTFTVATPPSNGTVVVNPDD187 pKa = 2.68GTYY190 pKa = 10.12TYY192 pKa = 10.56TPAPNYY198 pKa = 10.61NGTDD202 pKa = 3.31SFTVTVSDD210 pKa = 3.99GKK212 pKa = 11.28GGTTTVTVNVNVTPVNDD229 pKa = 3.77APVATSPAIVTNKK242 pKa = 9.94NIPVNGTITASDD254 pKa = 3.54VDD256 pKa = 4.0GDD258 pKa = 4.09ALTFTVSTPPAHH270 pKa = 5.76GTVVVNADD278 pKa = 3.0GTYY281 pKa = 10.14TYY283 pKa = 10.48TPANNYY289 pKa = 9.78SGSDD293 pKa = 3.45VFTVTVSDD301 pKa = 3.96GKK303 pKa = 11.28GGTTTVTINVTVNPTNVAPVVTAPAVTGNEE333 pKa = 3.95DD334 pKa = 3.45SPINGTITATDD345 pKa = 3.62ADD347 pKa = 4.47GDD349 pKa = 4.2PLTFTVATPPANGTVVVNPDD369 pKa = 2.68GTYY372 pKa = 10.15TYY374 pKa = 10.66TPVPNYY380 pKa = 10.71NGTDD384 pKa = 3.37SFTVTVSDD392 pKa = 3.99GKK394 pKa = 11.28GGTTTVTINVTVTPVNDD411 pKa = 3.82APVATAPAIVTNKK424 pKa = 9.91NIPVNGTITASDD436 pKa = 3.54VDD438 pKa = 4.0GDD440 pKa = 4.09ALTFTVSTPPAHH452 pKa = 5.76GTVVVNADD460 pKa = 3.0GTYY463 pKa = 10.14TYY465 pKa = 10.48TPANNYY471 pKa = 9.78SGSDD475 pKa = 3.45VFTVTVSDD483 pKa = 3.96GKK485 pKa = 11.28GGTTTVTINVTINPTNVAPVVTAPAVTGNEE515 pKa = 3.95DD516 pKa = 3.45SPINGTITATDD527 pKa = 3.62ADD529 pKa = 4.47GDD531 pKa = 4.22PLTFTVSTPPAHH543 pKa = 5.66GTVVINPDD551 pKa = 2.67GTYY554 pKa = 10.01TYY556 pKa = 10.56TPAPNYY562 pKa = 10.61NGTDD566 pKa = 3.31SFTVTVSDD574 pKa = 3.99GKK576 pKa = 11.28GGTTTVTINVTVTPPEE592 pKa = 4.59TISXTPPAHH601 pKa = 5.29GTVVINPDD609 pKa = 2.67GTYY612 pKa = 10.01TYY614 pKa = 10.56TPAPNYY620 pKa = 10.61NGTDD624 pKa = 3.31SFTVTVSDD632 pKa = 3.99GKK634 pKa = 11.28GGTTTVTINVIINPVNDD651 pKa = 3.98APVATAPAIVTNKK664 pKa = 9.91NIPVNGTITASDD676 pKa = 3.54VDD678 pKa = 4.0GDD680 pKa = 4.09ALTFTVSTPPAHH692 pKa = 6.07GTVTVNADD700 pKa = 2.98GTYY703 pKa = 10.12TYY705 pKa = 10.48TPANNYY711 pKa = 9.78SGSDD715 pKa = 3.45VFTVTVSDD723 pKa = 4.07GKK725 pKa = 11.22GGTATVTINVTINPTNVAPVVTAPAVTGNEE755 pKa = 3.95DD756 pKa = 3.45SPINGTITATDD767 pKa = 3.62ADD769 pKa = 4.47GDD771 pKa = 4.2PLTFTVATPPAHH783 pKa = 5.67GTVTVNADD791 pKa = 2.98GTYY794 pKa = 9.93TYY796 pKa = 10.65TPAPNYY802 pKa = 10.61NGTDD806 pKa = 3.31SFTVTVSDD814 pKa = 3.99GKK816 pKa = 11.28GGTTTVTVNVNVTPVNDD833 pKa = 3.77APVATSPAIVTNKK846 pKa = 9.94NIPVNGTITASDD858 pKa = 3.54VDD860 pKa = 4.0GDD862 pKa = 4.09ALTFTVSTPPAHH874 pKa = 6.07GTVTVNADD882 pKa = 2.98GTYY885 pKa = 10.12TYY887 pKa = 10.48TPANNYY893 pKa = 9.78SGSDD897 pKa = 3.45VFTVTVSDD905 pKa = 3.96GKK907 pKa = 11.28GGTTTVTINVTVNPTNVAPFVTSPAVTGNEE937 pKa = 3.88DD938 pKa = 3.46SPINSSITATDD949 pKa = 3.53ADD951 pKa = 4.4GDD953 pKa = 4.22PLTFTVSTPPAHH965 pKa = 5.83GTVVVNPDD973 pKa = 2.68GTYY976 pKa = 10.12TYY978 pKa = 10.56TPAPNYY984 pKa = 10.61NGTDD988 pKa = 3.31SFTVTVSDD996 pKa = 3.99GKK998 pKa = 11.28GGTTTVTVNVTVTPVNDD1015 pKa = 3.61APVATSPAIVTNKK1028 pKa = 9.94NIPVNGTITASDD1040 pKa = 3.54VDD1042 pKa = 4.0GDD1044 pKa = 4.09ALTFTVSTPPAHH1056 pKa = 5.91GTVTVKK1062 pKa = 10.95ADD1064 pKa = 3.24GTYY1067 pKa = 10.18TYY1069 pKa = 10.32TPANNYY1075 pKa = 9.78SGSDD1079 pKa = 3.45VFTVTVSDD1087 pKa = 3.96GKK1089 pKa = 11.28GGTTTVTINVTINPTNVAPVVTAPAVTGNEE1119 pKa = 3.95DD1120 pKa = 3.45SPINGTITATDD1131 pKa = 3.6ADD1133 pKa = 4.51GDD1135 pKa = 4.12PLIFTVATPPAHH1147 pKa = 5.67GTVTVNADD1155 pKa = 3.02GTYY1158 pKa = 10.07TYY1160 pKa = 10.69IPAPNYY1166 pKa = 10.61NGTDD1170 pKa = 3.31SFTVTVSDD1178 pKa = 3.99GKK1180 pKa = 11.28GGTTTVTVNVTVTPVNDD1197 pKa = 3.91APVTTAPAVTTPQNTAVNGAITASDD1222 pKa = 3.54VDD1224 pKa = 3.93GDD1226 pKa = 4.21ALTFTLTAQPAHH1238 pKa = 5.74GTVVVNANGTYY1249 pKa = 9.83TYY1251 pKa = 9.89TPTAGYY1257 pKa = 9.76SGNDD1261 pKa = 3.09TFTITVSDD1269 pKa = 3.94GKK1271 pKa = 10.97GGTVTVTVPVTVTLVAAPAMSLTKK1295 pKa = 9.92TATNTVSKK1303 pKa = 10.81VGDD1306 pKa = 3.61VINYY1310 pKa = 9.36NIIVTNTGNVTLTNVAVTDD1329 pKa = 4.21AGADD1333 pKa = 3.51AGSLTPAGIASLLPGSSVTVTARR1356 pKa = 11.84HH1357 pKa = 5.22TVTLTEE1363 pKa = 4.14VNAGSFSNQASATAQTPGGSILNKK1387 pKa = 9.85PKK1389 pKa = 10.6SDD1391 pKa = 4.18DD1392 pKa = 3.85PNTIAVDD1399 pKa = 3.75DD1400 pKa = 4.06ATVTVIAPASMITLVKK1416 pKa = 9.58TGTVSTDD1423 pKa = 3.12GNSITYY1429 pKa = 9.97SFSVKK1434 pKa = 8.97NTGNVTLHH1442 pKa = 7.06IITLVDD1448 pKa = 3.54AKK1450 pKa = 10.93LGLNRR1455 pKa = 11.84VIPGTLAPGATVTDD1469 pKa = 3.86SYY1471 pKa = 11.75VYY1473 pKa = 10.74QLTQADD1479 pKa = 3.97KK1480 pKa = 11.31DD1481 pKa = 3.84AGSVTNTASITGQTPANVAVSDD1503 pKa = 3.65ISGTAEE1509 pKa = 3.86NNNTPTVTLIPNNGSVALVKK1529 pKa = 10.16TSVFNGNKK1537 pKa = 6.32VTYY1540 pKa = 9.59TFTIKK1545 pKa = 9.56NTGTITLNTITLTDD1559 pKa = 3.69AKK1561 pKa = 10.69LGLNNKK1567 pKa = 9.2VITVAGGLAPGATTTDD1583 pKa = 3.41VEE1585 pKa = 5.23VYY1587 pKa = 10.12TLTQADD1593 pKa = 4.13KK1594 pKa = 11.53DD1595 pKa = 4.02LGTVTNTATVNARR1608 pKa = 11.84TLAGASVTDD1617 pKa = 3.32VSGTAEE1623 pKa = 4.27TNNTPTVTTFPKK1635 pKa = 10.65SPTAVDD1641 pKa = 4.71DD1642 pKa = 4.12KK1643 pKa = 11.36AQTVANSPVVISVLNNDD1660 pKa = 4.15DD1661 pKa = 4.74PGNSTLDD1668 pKa = 3.41KK1669 pKa = 10.54LTVEE1673 pKa = 4.55VVNAPKK1679 pKa = 10.54HH1680 pKa = 4.79GTVKK1684 pKa = 10.49VNADD1688 pKa = 3.28GTVTYY1693 pKa = 9.57TPDD1696 pKa = 3.5PGYY1699 pKa = 11.22VGDD1702 pKa = 5.2DD1703 pKa = 3.23VFTYY1707 pKa = 10.06RR1708 pKa = 11.84VKK1710 pKa = 10.74DD1711 pKa = 3.42AYY1713 pKa = 10.55GYY1715 pKa = 6.52YY1716 pKa = 10.08TNVATVTLTANFTGITVPNLFTPNGDD1742 pKa = 3.95GINDD1746 pKa = 3.63TFEE1749 pKa = 5.85IIGLNQYY1756 pKa = 8.03QVNEE1760 pKa = 3.97LQIVNRR1766 pKa = 11.84WGNEE1770 pKa = 3.45VFHH1773 pKa = 7.28AKK1775 pKa = 10.2GYY1777 pKa = 8.02QNNWTGEE1784 pKa = 3.99GLNEE1788 pKa = 3.67GTYY1791 pKa = 10.78YY1792 pKa = 11.08YY1793 pKa = 11.2LLRR1796 pKa = 11.84VKK1798 pKa = 10.45KK1799 pKa = 10.38AGSDD1803 pKa = 3.25QYY1805 pKa = 11.61EE1806 pKa = 4.23VFKK1809 pKa = 11.09GYY1811 pKa = 8.32ITLIKK1816 pKa = 10.31AFKK1819 pKa = 10.15KK1820 pKa = 10.59
Molecular weight: 184.11 kDa
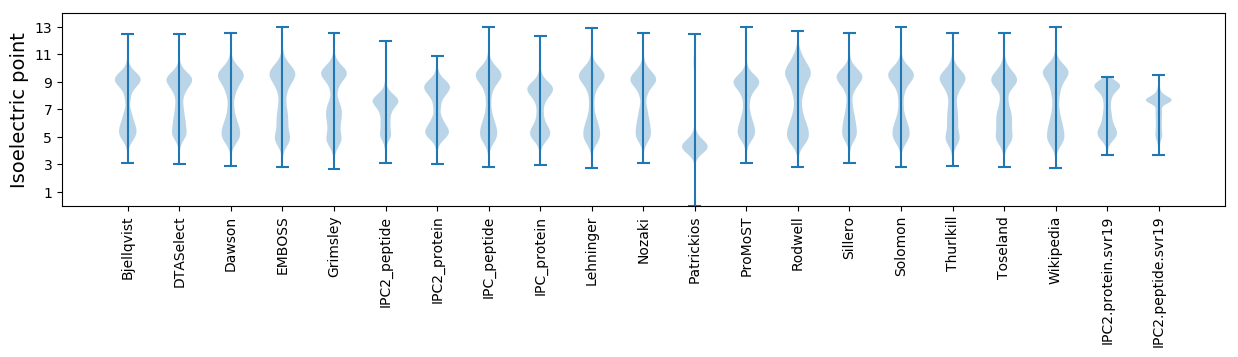
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q4GTF2|A0A0Q4GTF2_9SPHI Aspartate aminotransferase OS=Pedobacter sp. Leaf216 OX=1735684 GN=ASE74_19645 PE=4 SV=1
MM1 pKa = 7.65GIIIKK6 pKa = 10.49SGGIIIKK13 pKa = 10.35SRR15 pKa = 11.84GIIIKK20 pKa = 10.42SGGIIIKK27 pKa = 10.37SGGVIIKK34 pKa = 10.41SGGIIIKK41 pKa = 10.42SGGIIIKK48 pKa = 10.35SRR50 pKa = 11.84GIIIKK55 pKa = 10.37SGGVIIKK62 pKa = 10.41SGGIIIKK69 pKa = 10.42SGGIIIKK76 pKa = 10.42SGGIIIKK83 pKa = 10.42SGGIIIKK90 pKa = 10.35SRR92 pKa = 11.84GIIIKK97 pKa = 10.29SRR99 pKa = 11.84ASLL102 pKa = 3.49
MM1 pKa = 7.65GIIIKK6 pKa = 10.49SGGIIIKK13 pKa = 10.35SRR15 pKa = 11.84GIIIKK20 pKa = 10.42SGGIIIKK27 pKa = 10.37SGGVIIKK34 pKa = 10.41SGGIIIKK41 pKa = 10.42SGGIIIKK48 pKa = 10.35SRR50 pKa = 11.84GIIIKK55 pKa = 10.37SGGVIIKK62 pKa = 10.41SGGIIIKK69 pKa = 10.42SGGIIIKK76 pKa = 10.42SGGIIIKK83 pKa = 10.42SGGIIIKK90 pKa = 10.35SRR92 pKa = 11.84GIIIKK97 pKa = 10.29SRR99 pKa = 11.84ASLL102 pKa = 3.49
Molecular weight: 10.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1606351 |
36 |
4486 |
339.6 |
38.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.537 ± 0.036 | 0.729 ± 0.011 |
5.351 ± 0.025 | 5.598 ± 0.031 |
5.057 ± 0.03 | 6.806 ± 0.038 |
1.732 ± 0.017 | 7.493 ± 0.033 |
7.594 ± 0.042 | 9.401 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.151 ± 0.016 | 6.073 ± 0.034 |
3.679 ± 0.022 | 3.566 ± 0.023 |
3.553 ± 0.024 | 6.337 ± 0.029 |
5.881 ± 0.066 | 6.163 ± 0.029 |
1.162 ± 0.015 | 4.134 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
