
Marinobacter sp. AC-23
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter; unclassified Marinobacter
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
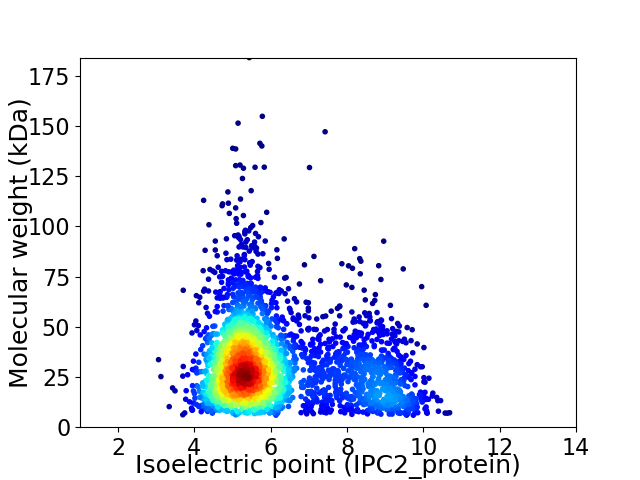
Virtual 2D-PAGE plot for 3073 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S2CDY8|A0A1S2CDY8_9ALTE Uncharacterized protein OS=Marinobacter sp. AC-23 OX=1879031 GN=BCA33_02885 PE=4 SV=1
MM1 pKa = 7.3KK2 pKa = 10.08LKK4 pKa = 10.45YY5 pKa = 9.73SAAIICLALSPALVSAEE22 pKa = 4.05PEE24 pKa = 4.05AGDD27 pKa = 4.41GLFALTGTGSSDD39 pKa = 3.52NEE41 pKa = 3.78FDD43 pKa = 3.85NNVAAVSFDD52 pKa = 3.28VGKK55 pKa = 10.54FFSDD59 pKa = 2.99NLAAGLRR66 pKa = 11.84QSFGFADD73 pKa = 3.88SEE75 pKa = 4.72NNSSWSGASRR85 pKa = 11.84GFVDD89 pKa = 3.01YY90 pKa = 11.12HH91 pKa = 7.2FDD93 pKa = 3.65GGSWQPFVGANLGGIYY109 pKa = 10.38GDD111 pKa = 4.96DD112 pKa = 3.41VDD114 pKa = 4.06EE115 pKa = 4.62TFFAGPEE122 pKa = 4.04AGVKK126 pKa = 10.15YY127 pKa = 8.99YY128 pKa = 10.75VRR130 pKa = 11.84EE131 pKa = 3.8KK132 pKa = 10.52TYY134 pKa = 9.25ITVQMEE140 pKa = 4.12YY141 pKa = 10.58QVFFDD146 pKa = 4.23SASDD150 pKa = 3.69ADD152 pKa = 4.99DD153 pKa = 4.61NFDD156 pKa = 4.77DD157 pKa = 5.12GAFAYY162 pKa = 10.43SAGIGYY168 pKa = 9.17NFF170 pKa = 4.18
MM1 pKa = 7.3KK2 pKa = 10.08LKK4 pKa = 10.45YY5 pKa = 9.73SAAIICLALSPALVSAEE22 pKa = 4.05PEE24 pKa = 4.05AGDD27 pKa = 4.41GLFALTGTGSSDD39 pKa = 3.52NEE41 pKa = 3.78FDD43 pKa = 3.85NNVAAVSFDD52 pKa = 3.28VGKK55 pKa = 10.54FFSDD59 pKa = 2.99NLAAGLRR66 pKa = 11.84QSFGFADD73 pKa = 3.88SEE75 pKa = 4.72NNSSWSGASRR85 pKa = 11.84GFVDD89 pKa = 3.01YY90 pKa = 11.12HH91 pKa = 7.2FDD93 pKa = 3.65GGSWQPFVGANLGGIYY109 pKa = 10.38GDD111 pKa = 4.96DD112 pKa = 3.41VDD114 pKa = 4.06EE115 pKa = 4.62TFFAGPEE122 pKa = 4.04AGVKK126 pKa = 10.15YY127 pKa = 8.99YY128 pKa = 10.75VRR130 pKa = 11.84EE131 pKa = 3.8KK132 pKa = 10.52TYY134 pKa = 9.25ITVQMEE140 pKa = 4.12YY141 pKa = 10.58QVFFDD146 pKa = 4.23SASDD150 pKa = 3.69ADD152 pKa = 4.99DD153 pKa = 4.61NFDD156 pKa = 4.77DD157 pKa = 5.12GAFAYY162 pKa = 10.43SAGIGYY168 pKa = 9.17NFF170 pKa = 4.18
Molecular weight: 18.18 kDa
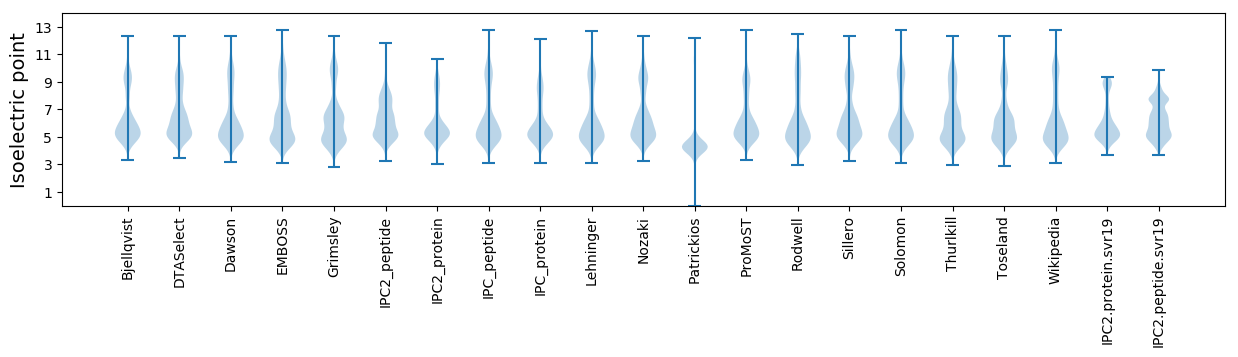
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S2CE99|A0A1S2CE99_9ALTE 2-polyprenyl-3-methyl-5-hydroxy-6-metoxy-1 4-benzoquinol methylase OS=Marinobacter sp. AC-23 OX=1879031 GN=BCA33_02805 PE=4 SV=1
MM1 pKa = 7.35NVTTIGIDD9 pKa = 3.26LAKK12 pKa = 9.84TVFSIHH18 pKa = 6.11GTNQHH23 pKa = 5.19GKK25 pKa = 8.42VVVRR29 pKa = 11.84KK30 pKa = 9.72RR31 pKa = 11.84LNRR34 pKa = 11.84PKK36 pKa = 10.58LLAFFAQLPPCLIGMEE52 pKa = 4.34ACSGAHH58 pKa = 4.01YY59 pKa = 8.21WARR62 pKa = 11.84EE63 pKa = 4.0LTKK66 pKa = 10.39LGHH69 pKa = 6.81DD70 pKa = 3.89ARR72 pKa = 11.84IIAPRR77 pKa = 11.84FVAPYY82 pKa = 9.82RR83 pKa = 11.84KK84 pKa = 9.62SSKK87 pKa = 10.76NDD89 pKa = 3.34DD90 pKa = 3.64NDD92 pKa = 3.81AEE94 pKa = 4.76AICEE98 pKa = 4.09AVSRR102 pKa = 11.84PNMRR106 pKa = 11.84FVPIKK111 pKa = 9.78TQDD114 pKa = 3.32QQAALCLHH122 pKa = 7.63RR123 pKa = 11.84IRR125 pKa = 11.84RR126 pKa = 11.84GLIKK130 pKa = 10.46EE131 pKa = 3.74RR132 pKa = 11.84TAQINQIRR140 pKa = 11.84GLLAEE145 pKa = 5.25FGLIIPQGRR154 pKa = 11.84YY155 pKa = 7.89HH156 pKa = 6.58ARR158 pKa = 11.84HH159 pKa = 6.91RR160 pKa = 11.84IPEE163 pKa = 4.03ILEE166 pKa = 4.14DD167 pKa = 4.23AEE169 pKa = 4.71NGLPMLARR177 pKa = 11.84RR178 pKa = 11.84LLNNINQRR186 pKa = 11.84ILQLDD191 pKa = 4.24EE192 pKa = 5.66EE193 pKa = 4.57ILTYY197 pKa = 10.67DD198 pKa = 4.33RR199 pKa = 11.84EE200 pKa = 4.37IEE202 pKa = 4.17SMARR206 pKa = 11.84RR207 pKa = 11.84DD208 pKa = 3.54EE209 pKa = 4.08RR210 pKa = 11.84AKK212 pKa = 10.82RR213 pKa = 11.84LMAIPGIGPQTATAILASAPDD234 pKa = 3.56PRR236 pKa = 11.84QFKK239 pKa = 10.21SGRR242 pKa = 11.84HH243 pKa = 3.6FAAWLGLVPRR253 pKa = 11.84QYY255 pKa = 10.49STGGKK260 pKa = 8.48PRR262 pKa = 11.84LGRR265 pKa = 11.84ITKK268 pKa = 10.47RR269 pKa = 11.84GDD271 pKa = 2.81KK272 pKa = 10.35YY273 pKa = 11.32LRR275 pKa = 11.84TLLIHH280 pKa = 5.89GTRR283 pKa = 11.84AVIAKK288 pKa = 9.85LADD291 pKa = 3.48KK292 pKa = 10.75NDD294 pKa = 4.06RR295 pKa = 11.84LSGWARR301 pKa = 11.84EE302 pKa = 4.02LVEE305 pKa = 3.64RR306 pKa = 11.84RR307 pKa = 11.84GYY309 pKa = 10.24KK310 pKa = 9.6RR311 pKa = 11.84AAVALAAKK319 pKa = 9.37NARR322 pKa = 11.84IIWRR326 pKa = 11.84YY327 pKa = 8.61
MM1 pKa = 7.35NVTTIGIDD9 pKa = 3.26LAKK12 pKa = 9.84TVFSIHH18 pKa = 6.11GTNQHH23 pKa = 5.19GKK25 pKa = 8.42VVVRR29 pKa = 11.84KK30 pKa = 9.72RR31 pKa = 11.84LNRR34 pKa = 11.84PKK36 pKa = 10.58LLAFFAQLPPCLIGMEE52 pKa = 4.34ACSGAHH58 pKa = 4.01YY59 pKa = 8.21WARR62 pKa = 11.84EE63 pKa = 4.0LTKK66 pKa = 10.39LGHH69 pKa = 6.81DD70 pKa = 3.89ARR72 pKa = 11.84IIAPRR77 pKa = 11.84FVAPYY82 pKa = 9.82RR83 pKa = 11.84KK84 pKa = 9.62SSKK87 pKa = 10.76NDD89 pKa = 3.34DD90 pKa = 3.64NDD92 pKa = 3.81AEE94 pKa = 4.76AICEE98 pKa = 4.09AVSRR102 pKa = 11.84PNMRR106 pKa = 11.84FVPIKK111 pKa = 9.78TQDD114 pKa = 3.32QQAALCLHH122 pKa = 7.63RR123 pKa = 11.84IRR125 pKa = 11.84RR126 pKa = 11.84GLIKK130 pKa = 10.46EE131 pKa = 3.74RR132 pKa = 11.84TAQINQIRR140 pKa = 11.84GLLAEE145 pKa = 5.25FGLIIPQGRR154 pKa = 11.84YY155 pKa = 7.89HH156 pKa = 6.58ARR158 pKa = 11.84HH159 pKa = 6.91RR160 pKa = 11.84IPEE163 pKa = 4.03ILEE166 pKa = 4.14DD167 pKa = 4.23AEE169 pKa = 4.71NGLPMLARR177 pKa = 11.84RR178 pKa = 11.84LLNNINQRR186 pKa = 11.84ILQLDD191 pKa = 4.24EE192 pKa = 5.66EE193 pKa = 4.57ILTYY197 pKa = 10.67DD198 pKa = 4.33RR199 pKa = 11.84EE200 pKa = 4.37IEE202 pKa = 4.17SMARR206 pKa = 11.84RR207 pKa = 11.84DD208 pKa = 3.54EE209 pKa = 4.08RR210 pKa = 11.84AKK212 pKa = 10.82RR213 pKa = 11.84LMAIPGIGPQTATAILASAPDD234 pKa = 3.56PRR236 pKa = 11.84QFKK239 pKa = 10.21SGRR242 pKa = 11.84HH243 pKa = 3.6FAAWLGLVPRR253 pKa = 11.84QYY255 pKa = 10.49STGGKK260 pKa = 8.48PRR262 pKa = 11.84LGRR265 pKa = 11.84ITKK268 pKa = 10.47RR269 pKa = 11.84GDD271 pKa = 2.81KK272 pKa = 10.35YY273 pKa = 11.32LRR275 pKa = 11.84TLLIHH280 pKa = 5.89GTRR283 pKa = 11.84AVIAKK288 pKa = 9.85LADD291 pKa = 3.48KK292 pKa = 10.75NDD294 pKa = 4.06RR295 pKa = 11.84LSGWARR301 pKa = 11.84EE302 pKa = 4.02LVEE305 pKa = 3.64RR306 pKa = 11.84RR307 pKa = 11.84GYY309 pKa = 10.24KK310 pKa = 9.6RR311 pKa = 11.84AAVALAAKK319 pKa = 9.37NARR322 pKa = 11.84IIWRR326 pKa = 11.84YY327 pKa = 8.61
Molecular weight: 37.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
917237 |
51 |
1623 |
298.5 |
32.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.817 ± 0.05 | 0.945 ± 0.013 |
5.521 ± 0.039 | 6.432 ± 0.047 |
3.859 ± 0.028 | 7.844 ± 0.039 |
2.18 ± 0.021 | 5.42 ± 0.037 |
4.023 ± 0.037 | 10.609 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.663 ± 0.023 | 3.42 ± 0.028 |
4.58 ± 0.032 | 3.999 ± 0.033 |
6.109 ± 0.033 | 6.167 ± 0.037 |
5.192 ± 0.027 | 7.262 ± 0.04 |
1.373 ± 0.022 | 2.585 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
