
Actinobaculum suis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group;
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
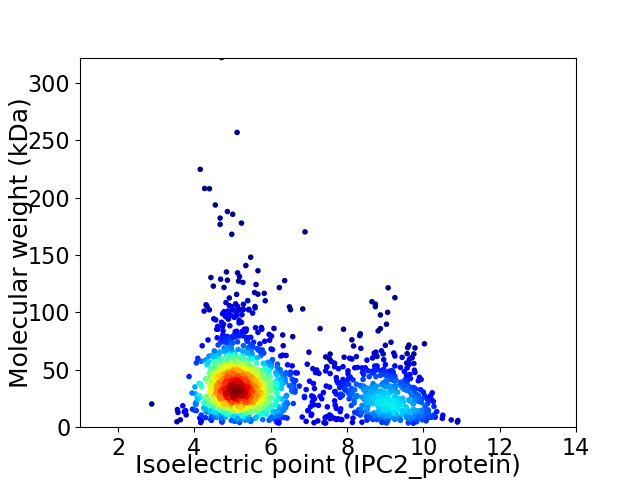
Virtual 2D-PAGE plot for 1723 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K9ET46|A0A0K9ET46_9ACTO HAD family hydrolase OS=Actinobaculum suis OX=1657 GN=nagD_1 PE=3 SV=1
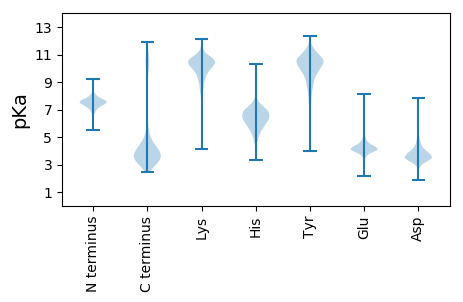
MM1 pKa = 7.14ATDD4 pKa = 3.65YY5 pKa = 10.33DD6 pKa = 3.7APRR9 pKa = 11.84NKK11 pKa = 10.59DD12 pKa = 2.83EE13 pKa = 4.24GLRR16 pKa = 11.84EE17 pKa = 3.91EE18 pKa = 5.16SLQQLTSQSDD28 pKa = 3.31QSSNVDD34 pKa = 3.19EE35 pKa = 5.54DD36 pKa = 3.69EE37 pKa = 4.45VEE39 pKa = 3.91AAEE42 pKa = 4.66GFEE45 pKa = 5.34LPGADD50 pKa = 3.82LSNVEE55 pKa = 4.47LSVNVIPAQDD65 pKa = 3.94DD66 pKa = 3.81EE67 pKa = 5.06FTCSVCFLVHH77 pKa = 6.54HH78 pKa = 7.0RR79 pKa = 11.84SQLDD83 pKa = 3.3HH84 pKa = 6.86MEE86 pKa = 5.38DD87 pKa = 3.72GLPVCMDD94 pKa = 3.96CAFF97 pKa = 4.05
MM1 pKa = 7.14ATDD4 pKa = 3.65YY5 pKa = 10.33DD6 pKa = 3.7APRR9 pKa = 11.84NKK11 pKa = 10.59DD12 pKa = 2.83EE13 pKa = 4.24GLRR16 pKa = 11.84EE17 pKa = 3.91EE18 pKa = 5.16SLQQLTSQSDD28 pKa = 3.31QSSNVDD34 pKa = 3.19EE35 pKa = 5.54DD36 pKa = 3.69EE37 pKa = 4.45VEE39 pKa = 3.91AAEE42 pKa = 4.66GFEE45 pKa = 5.34LPGADD50 pKa = 3.82LSNVEE55 pKa = 4.47LSVNVIPAQDD65 pKa = 3.94DD66 pKa = 3.81EE67 pKa = 5.06FTCSVCFLVHH77 pKa = 6.54HH78 pKa = 7.0RR79 pKa = 11.84SQLDD83 pKa = 3.3HH84 pKa = 6.86MEE86 pKa = 5.38DD87 pKa = 3.72GLPVCMDD94 pKa = 3.96CAFF97 pKa = 4.05
Molecular weight: 10.74 kDa
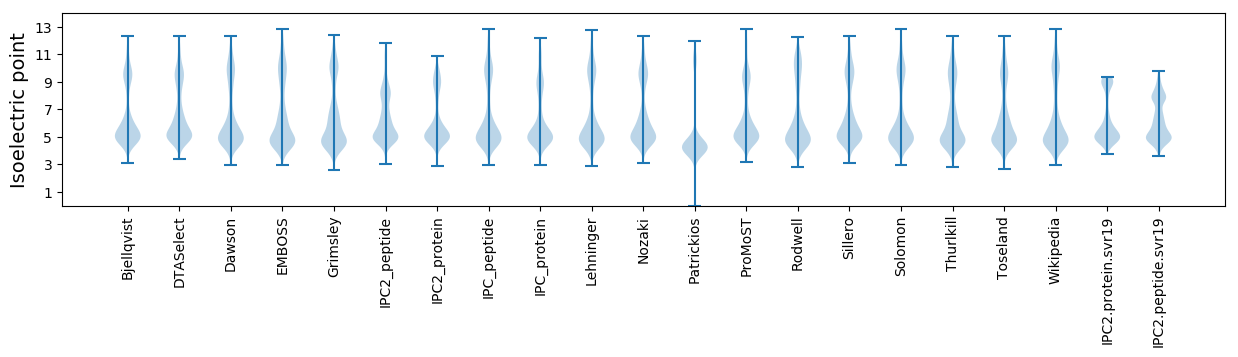
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6ZBX4|A0A1G6ZBX4_9ACTO UDP-N-acetylmuramate--L-alanine ligase OS=Actinobaculum suis OX=1657 GN=murC PE=3 SV=1
MM1 pKa = 7.67AKK3 pKa = 9.67TGGTHH8 pKa = 7.67DD9 pKa = 3.19AATRR13 pKa = 11.84HH14 pKa = 6.01DD15 pKa = 4.08APTRR19 pKa = 11.84ATRR22 pKa = 11.84RR23 pKa = 11.84GSRR26 pKa = 11.84GATGQARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84TPKK38 pKa = 10.31GKK40 pKa = 9.18PVTGATRR47 pKa = 11.84LPAEE51 pKa = 4.24NAEE54 pKa = 4.25KK55 pKa = 10.75VRR57 pKa = 11.84DD58 pKa = 3.96VMRR61 pKa = 11.84HH62 pKa = 5.15SIIMLAVATLVLTVLGLVMVFSAVTPGAMRR92 pKa = 11.84SAAAGATNTFRR103 pKa = 11.84GVYY106 pKa = 9.67LQIFYY111 pKa = 10.49ACVGIVVAAILSRR124 pKa = 11.84FPIEE128 pKa = 4.18FFRR131 pKa = 11.84KK132 pKa = 8.56SWAAFWAGALLLQCLVFSPLGVAVAGNRR160 pKa = 11.84NWVALGPIRR169 pKa = 11.84MQPSEE174 pKa = 4.23FLKK177 pKa = 10.78LATIIALAATLVNVRR192 pKa = 11.84GAASVNLLRR201 pKa = 11.84RR202 pKa = 11.84EE203 pKa = 3.68EE204 pKa = 4.11WEE206 pKa = 5.29DD207 pKa = 2.92IKK209 pKa = 11.59NWVWPIAIFLLSAGLVGLGHH229 pKa = 7.65DD230 pKa = 3.87MGTVLVFALFTAGMLWLAGMAWQWFAVGGLAGILGTALAIAVSLSRR276 pKa = 11.84LARR279 pKa = 11.84VQDD282 pKa = 3.58FFEE285 pKa = 5.53NIFTLPAGGSPTQADD300 pKa = 3.76YY301 pKa = 11.71ALWAFGSGGLGGSGLGTGIEE321 pKa = 4.08KK322 pKa = 10.29WPGNLAEE329 pKa = 4.7AQTDD333 pKa = 4.09FIFAVVGEE341 pKa = 4.16EE342 pKa = 5.24LGFLGCAAVVILFLMVGISLVRR364 pKa = 11.84ICIHH368 pKa = 5.96HH369 pKa = 6.97PSRR372 pKa = 11.84FAKK375 pKa = 9.33LTVGGIALWLCGQALANMLVVTSMLPVFGVPLPFMSQGGSSVISCLMAVGVAISAAFSVEE435 pKa = 3.76GVRR438 pKa = 11.84ASFKK442 pKa = 10.42QRR444 pKa = 11.84PHH446 pKa = 5.1LVKK449 pKa = 10.14RR450 pKa = 11.84VVSIVSKK457 pKa = 9.7TRR459 pKa = 3.02
MM1 pKa = 7.67AKK3 pKa = 9.67TGGTHH8 pKa = 7.67DD9 pKa = 3.19AATRR13 pKa = 11.84HH14 pKa = 6.01DD15 pKa = 4.08APTRR19 pKa = 11.84ATRR22 pKa = 11.84RR23 pKa = 11.84GSRR26 pKa = 11.84GATGQARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84TPKK38 pKa = 10.31GKK40 pKa = 9.18PVTGATRR47 pKa = 11.84LPAEE51 pKa = 4.24NAEE54 pKa = 4.25KK55 pKa = 10.75VRR57 pKa = 11.84DD58 pKa = 3.96VMRR61 pKa = 11.84HH62 pKa = 5.15SIIMLAVATLVLTVLGLVMVFSAVTPGAMRR92 pKa = 11.84SAAAGATNTFRR103 pKa = 11.84GVYY106 pKa = 9.67LQIFYY111 pKa = 10.49ACVGIVVAAILSRR124 pKa = 11.84FPIEE128 pKa = 4.18FFRR131 pKa = 11.84KK132 pKa = 8.56SWAAFWAGALLLQCLVFSPLGVAVAGNRR160 pKa = 11.84NWVALGPIRR169 pKa = 11.84MQPSEE174 pKa = 4.23FLKK177 pKa = 10.78LATIIALAATLVNVRR192 pKa = 11.84GAASVNLLRR201 pKa = 11.84RR202 pKa = 11.84EE203 pKa = 3.68EE204 pKa = 4.11WEE206 pKa = 5.29DD207 pKa = 2.92IKK209 pKa = 11.59NWVWPIAIFLLSAGLVGLGHH229 pKa = 7.65DD230 pKa = 3.87MGTVLVFALFTAGMLWLAGMAWQWFAVGGLAGILGTALAIAVSLSRR276 pKa = 11.84LARR279 pKa = 11.84VQDD282 pKa = 3.58FFEE285 pKa = 5.53NIFTLPAGGSPTQADD300 pKa = 3.76YY301 pKa = 11.71ALWAFGSGGLGGSGLGTGIEE321 pKa = 4.08KK322 pKa = 10.29WPGNLAEE329 pKa = 4.7AQTDD333 pKa = 4.09FIFAVVGEE341 pKa = 4.16EE342 pKa = 5.24LGFLGCAAVVILFLMVGISLVRR364 pKa = 11.84ICIHH368 pKa = 5.96HH369 pKa = 6.97PSRR372 pKa = 11.84FAKK375 pKa = 9.33LTVGGIALWLCGQALANMLVVTSMLPVFGVPLPFMSQGGSSVISCLMAVGVAISAAFSVEE435 pKa = 3.76GVRR438 pKa = 11.84ASFKK442 pKa = 10.42QRR444 pKa = 11.84PHH446 pKa = 5.1LVKK449 pKa = 10.14RR450 pKa = 11.84VVSIVSKK457 pKa = 9.7TRR459 pKa = 3.02
Molecular weight: 48.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
622842 |
30 |
3033 |
361.5 |
38.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.706 ± 0.087 | 0.69 ± 0.014 |
5.154 ± 0.054 | 6.609 ± 0.054 |
3.135 ± 0.032 | 8.641 ± 0.057 |
1.889 ± 0.025 | 5.011 ± 0.044 |
3.211 ± 0.044 | 9.015 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.015 ± 0.027 | 2.911 ± 0.041 |
5.412 ± 0.048 | 3.632 ± 0.031 |
6.471 ± 0.065 | 5.802 ± 0.041 |
6.204 ± 0.047 | 7.691 ± 0.052 |
1.338 ± 0.023 | 2.464 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
