
Hirschia baltica (strain ATCC 49814 / DSM 5838 / IFAM 1418)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomonadales; Hyphomonadaceae; Hirschia; Hirschia baltica
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
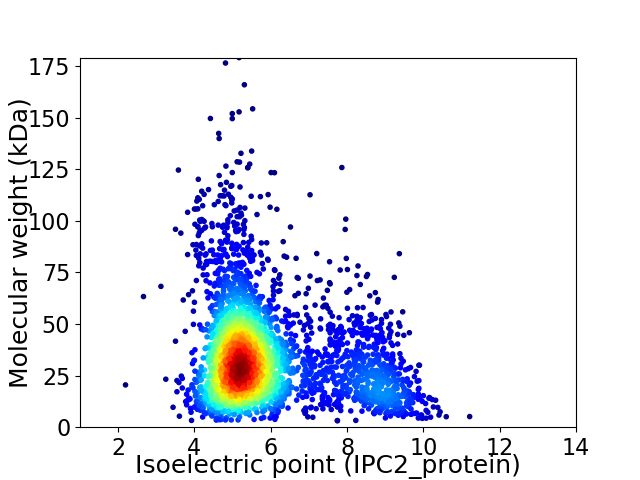
Virtual 2D-PAGE plot for 3187 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6XQF2|C6XQF2_HIRBI Uroporphyrinogen III methylase OS=Hirschia baltica (strain ATCC 49814 / DSM 5838 / IFAM 1418) OX=582402 GN=Hbal_2778 PE=3 SV=1
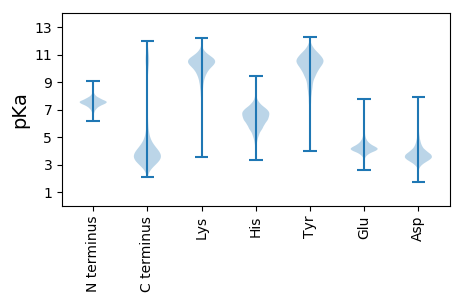
MM1 pKa = 7.58IMTAPAIFVRR11 pKa = 11.84KK12 pKa = 9.21NIKK15 pKa = 8.74RR16 pKa = 11.84TSPLVFLTILYY27 pKa = 7.52STCSPLALGQEE38 pKa = 4.32ISSATSTPVEE48 pKa = 4.37TATLDD53 pKa = 3.37NGSASDD59 pKa = 4.4LTITDD64 pKa = 4.3DD65 pKa = 6.26GSIEE69 pKa = 4.01WDD71 pKa = 3.58SVDD74 pKa = 3.72DD75 pKa = 3.98QVAVTVNSSNNVTIDD90 pKa = 3.6GDD92 pKa = 3.75ITLEE96 pKa = 4.06NSDD99 pKa = 3.85NNVGVKK105 pKa = 10.07INPNLSSTINLSGAINLIEE124 pKa = 5.14DD125 pKa = 3.95YY126 pKa = 11.35ARR128 pKa = 11.84TDD130 pKa = 3.15TDD132 pKa = 5.5DD133 pKa = 6.3DD134 pKa = 5.04DD135 pKa = 6.64DD136 pKa = 6.67LDD138 pKa = 3.99GLIAEE143 pKa = 4.62GTNRR147 pKa = 11.84HH148 pKa = 6.04GIFLEE153 pKa = 4.21SGGINTGDD161 pKa = 4.47IILQSGSSIYY171 pKa = 10.8VEE173 pKa = 4.88GNNSTAITLQSGLNGNYY190 pKa = 9.39IQDD193 pKa = 3.58GSISVYY199 pKa = 10.2GANSTALDD207 pKa = 3.37IQNDD211 pKa = 3.47VNGNVRR217 pKa = 11.84VSGGVTANGEE227 pKa = 4.0NASAISIAGDD237 pKa = 2.99ITGNLTLEE245 pKa = 4.11NTITSTGFTSTTSSNYY261 pKa = 9.69IGPDD265 pKa = 4.27DD266 pKa = 5.4IDD268 pKa = 6.07DD269 pKa = 4.1DD270 pKa = 4.25TAALEE275 pKa = 4.64DD276 pKa = 4.46RR277 pKa = 11.84LDD279 pKa = 3.94NDD281 pKa = 4.78DD282 pKa = 4.98LFNSASSVSIGGSIANGLLINGYY305 pKa = 9.62HH306 pKa = 7.46DD307 pKa = 4.03YY308 pKa = 11.29VDD310 pKa = 5.1DD311 pKa = 5.85GIAVDD316 pKa = 5.33DD317 pKa = 5.51DD318 pKa = 3.99EE319 pKa = 5.06TKK321 pKa = 10.44DD322 pKa = 3.82TIDD325 pKa = 4.86DD326 pKa = 3.87YY327 pKa = 11.98NEE329 pKa = 3.77NRR331 pKa = 11.84GTGYY335 pKa = 10.11ISTYY339 pKa = 10.06GSSPALQITPDD350 pKa = 4.51LIIGSSSDD358 pKa = 3.52LVLGSVVEE366 pKa = 4.45TVRR369 pKa = 11.84DD370 pKa = 3.74TLDD373 pKa = 3.79DD374 pKa = 5.81DD375 pKa = 4.6EE376 pKa = 7.45DD377 pKa = 4.48EE378 pKa = 4.75DD379 pKa = 4.55TNEE382 pKa = 3.8VLATFNFDD390 pKa = 3.05QGLVNLGTIYY400 pKa = 11.14ADD402 pKa = 3.53GQNVGFSATALEE414 pKa = 4.49ISGSADD420 pKa = 3.01GNLNTIITNGILNSGTMSTTAFEE443 pKa = 4.92ANATTVNLGQGAIISSITNDD463 pKa = 2.85GSIISNSTGSSGSNSIAIQISNGANLEE490 pKa = 4.38TIEE493 pKa = 3.97NTGYY497 pKa = 8.44ITAKK501 pKa = 9.06ITDD504 pKa = 3.69DD505 pKa = 3.52TGYY508 pKa = 8.59STAILDD514 pKa = 3.71HH515 pKa = 6.88SGTLTSIFNNGYY527 pKa = 8.82ISSYY531 pKa = 10.96YY532 pKa = 10.47VDD534 pKa = 5.57DD535 pKa = 6.09DD536 pKa = 5.33SDD538 pKa = 4.35DD539 pKa = 5.42DD540 pKa = 5.45EE541 pKa = 7.11YY542 pKa = 11.77DD543 pKa = 4.16DD544 pKa = 4.66NDD546 pKa = 3.5NKK548 pKa = 10.39ISEE551 pKa = 4.32FTGRR555 pKa = 11.84SIAIDD560 pKa = 3.76VSQHH564 pKa = 4.68TASQGVTIVQGNTNSTSPSIYY585 pKa = 10.61GDD587 pKa = 3.29ILFGAGNDD595 pKa = 3.69TLDD598 pKa = 3.74IQDD601 pKa = 3.65GTVAGDD607 pKa = 3.52TYY609 pKa = 10.93FGSGTANFYY618 pKa = 11.07LSDD621 pKa = 3.29ATYY624 pKa = 9.58TGDD627 pKa = 3.23VHH629 pKa = 7.81FEE631 pKa = 4.04SEE633 pKa = 4.25GFNFQSTQGQYY644 pKa = 10.53FGNLNFYY651 pKa = 9.93NNSGSAVFQNNSYY664 pKa = 10.76FSGSLLDD671 pKa = 4.27SDD673 pKa = 5.41NIDD676 pKa = 5.0LIIDD680 pKa = 4.03TSTIYY685 pKa = 10.19ISQDD689 pKa = 2.91SPITLNSLNITGASTLGVIISNSEE713 pKa = 4.36HH714 pKa = 7.0IDD716 pKa = 3.42TPYY719 pKa = 9.4ITATNTAIVGDD730 pKa = 4.48SVTLSSQFTNFVGGDD745 pKa = 3.57FSRR748 pKa = 11.84TILSAPSLDD757 pKa = 3.33VDD759 pKa = 3.39IDD761 pKa = 4.29SISFDD766 pKa = 3.41TSGLSWLYY774 pKa = 9.11NAEE777 pKa = 4.3YY778 pKa = 10.45ILDD781 pKa = 3.78QEE783 pKa = 4.6ADD785 pKa = 3.76GNQSISLQFSQKK797 pKa = 8.82TSEE800 pKa = 4.47DD801 pKa = 3.59LGLDD805 pKa = 3.47AQQDD809 pKa = 3.5RR810 pKa = 11.84TFASFIEE817 pKa = 4.71MISNQQNAGSEE828 pKa = 4.39FVKK831 pKa = 9.76ITDD834 pKa = 3.21EE835 pKa = 4.43TTFLEE840 pKa = 4.89AYY842 pKa = 10.0EE843 pKa = 4.8SILPQHH849 pKa = 6.73SDD851 pKa = 2.79VALRR855 pKa = 11.84HH856 pKa = 6.02LDD858 pKa = 3.16AHH860 pKa = 6.23TNTLNSMVSEE870 pKa = 4.34RR871 pKa = 11.84MALLRR876 pKa = 11.84DD877 pKa = 3.69SSNTSNGFWLQEE889 pKa = 4.11LVSQTTVDD897 pKa = 2.98ASEE900 pKa = 4.15NVNAYY905 pKa = 9.77SGRR908 pKa = 11.84GFGFTAGVDD917 pKa = 3.5RR918 pKa = 11.84RR919 pKa = 11.84IGFIDD924 pKa = 3.56SMGLTFSINDD934 pKa = 3.49EE935 pKa = 4.29KK936 pKa = 11.76YY937 pKa = 7.67EE938 pKa = 4.26TTTSAFNEE946 pKa = 4.27TSSTNFALGFYY957 pKa = 9.33IAEE960 pKa = 4.5RR961 pKa = 11.84IGFADD966 pKa = 4.11LQMSAQIGQTNFKK979 pKa = 10.24SEE981 pKa = 4.16RR982 pKa = 11.84EE983 pKa = 4.17VNFEE987 pKa = 4.18GFNSDD992 pKa = 5.54IEE994 pKa = 4.58GDD996 pKa = 3.49WSGLTQAYY1004 pKa = 6.97STQISSPQKK1013 pKa = 10.32LGWLRR1018 pKa = 11.84VTPHH1022 pKa = 6.64IGASYY1027 pKa = 10.79ISLNQDD1033 pKa = 2.65AYY1035 pKa = 10.7EE1036 pKa = 4.12EE1037 pKa = 4.25TASNGFNLAFSEE1049 pKa = 4.53SEE1051 pKa = 4.43SNKK1054 pKa = 8.8LTGSAGVSLGVFWPSNSGRR1073 pKa = 11.84NSFDD1077 pKa = 3.22LSEE1080 pKa = 4.25NQQSNSSLNGWHH1092 pKa = 6.76AAVDD1096 pKa = 3.66LGIRR1100 pKa = 11.84DD1101 pKa = 4.1TLSSTNYY1108 pKa = 9.74DD1109 pKa = 2.87AVANFVGYY1117 pKa = 10.73DD1118 pKa = 3.36SAFNVYY1124 pKa = 10.16SDD1126 pKa = 4.83EE1127 pKa = 4.48IFGQAFSTGISLVGIGDD1144 pKa = 4.06FASFRR1149 pKa = 11.84LSANAEE1155 pKa = 4.04LSEE1158 pKa = 4.19GASIYY1163 pKa = 10.86SGAASLRR1170 pKa = 11.84FKK1172 pKa = 10.96FF1173 pKa = 4.26
MM1 pKa = 7.58IMTAPAIFVRR11 pKa = 11.84KK12 pKa = 9.21NIKK15 pKa = 8.74RR16 pKa = 11.84TSPLVFLTILYY27 pKa = 7.52STCSPLALGQEE38 pKa = 4.32ISSATSTPVEE48 pKa = 4.37TATLDD53 pKa = 3.37NGSASDD59 pKa = 4.4LTITDD64 pKa = 4.3DD65 pKa = 6.26GSIEE69 pKa = 4.01WDD71 pKa = 3.58SVDD74 pKa = 3.72DD75 pKa = 3.98QVAVTVNSSNNVTIDD90 pKa = 3.6GDD92 pKa = 3.75ITLEE96 pKa = 4.06NSDD99 pKa = 3.85NNVGVKK105 pKa = 10.07INPNLSSTINLSGAINLIEE124 pKa = 5.14DD125 pKa = 3.95YY126 pKa = 11.35ARR128 pKa = 11.84TDD130 pKa = 3.15TDD132 pKa = 5.5DD133 pKa = 6.3DD134 pKa = 5.04DD135 pKa = 6.64DD136 pKa = 6.67LDD138 pKa = 3.99GLIAEE143 pKa = 4.62GTNRR147 pKa = 11.84HH148 pKa = 6.04GIFLEE153 pKa = 4.21SGGINTGDD161 pKa = 4.47IILQSGSSIYY171 pKa = 10.8VEE173 pKa = 4.88GNNSTAITLQSGLNGNYY190 pKa = 9.39IQDD193 pKa = 3.58GSISVYY199 pKa = 10.2GANSTALDD207 pKa = 3.37IQNDD211 pKa = 3.47VNGNVRR217 pKa = 11.84VSGGVTANGEE227 pKa = 4.0NASAISIAGDD237 pKa = 2.99ITGNLTLEE245 pKa = 4.11NTITSTGFTSTTSSNYY261 pKa = 9.69IGPDD265 pKa = 4.27DD266 pKa = 5.4IDD268 pKa = 6.07DD269 pKa = 4.1DD270 pKa = 4.25TAALEE275 pKa = 4.64DD276 pKa = 4.46RR277 pKa = 11.84LDD279 pKa = 3.94NDD281 pKa = 4.78DD282 pKa = 4.98LFNSASSVSIGGSIANGLLINGYY305 pKa = 9.62HH306 pKa = 7.46DD307 pKa = 4.03YY308 pKa = 11.29VDD310 pKa = 5.1DD311 pKa = 5.85GIAVDD316 pKa = 5.33DD317 pKa = 5.51DD318 pKa = 3.99EE319 pKa = 5.06TKK321 pKa = 10.44DD322 pKa = 3.82TIDD325 pKa = 4.86DD326 pKa = 3.87YY327 pKa = 11.98NEE329 pKa = 3.77NRR331 pKa = 11.84GTGYY335 pKa = 10.11ISTYY339 pKa = 10.06GSSPALQITPDD350 pKa = 4.51LIIGSSSDD358 pKa = 3.52LVLGSVVEE366 pKa = 4.45TVRR369 pKa = 11.84DD370 pKa = 3.74TLDD373 pKa = 3.79DD374 pKa = 5.81DD375 pKa = 4.6EE376 pKa = 7.45DD377 pKa = 4.48EE378 pKa = 4.75DD379 pKa = 4.55TNEE382 pKa = 3.8VLATFNFDD390 pKa = 3.05QGLVNLGTIYY400 pKa = 11.14ADD402 pKa = 3.53GQNVGFSATALEE414 pKa = 4.49ISGSADD420 pKa = 3.01GNLNTIITNGILNSGTMSTTAFEE443 pKa = 4.92ANATTVNLGQGAIISSITNDD463 pKa = 2.85GSIISNSTGSSGSNSIAIQISNGANLEE490 pKa = 4.38TIEE493 pKa = 3.97NTGYY497 pKa = 8.44ITAKK501 pKa = 9.06ITDD504 pKa = 3.69DD505 pKa = 3.52TGYY508 pKa = 8.59STAILDD514 pKa = 3.71HH515 pKa = 6.88SGTLTSIFNNGYY527 pKa = 8.82ISSYY531 pKa = 10.96YY532 pKa = 10.47VDD534 pKa = 5.57DD535 pKa = 6.09DD536 pKa = 5.33SDD538 pKa = 4.35DD539 pKa = 5.42DD540 pKa = 5.45EE541 pKa = 7.11YY542 pKa = 11.77DD543 pKa = 4.16DD544 pKa = 4.66NDD546 pKa = 3.5NKK548 pKa = 10.39ISEE551 pKa = 4.32FTGRR555 pKa = 11.84SIAIDD560 pKa = 3.76VSQHH564 pKa = 4.68TASQGVTIVQGNTNSTSPSIYY585 pKa = 10.61GDD587 pKa = 3.29ILFGAGNDD595 pKa = 3.69TLDD598 pKa = 3.74IQDD601 pKa = 3.65GTVAGDD607 pKa = 3.52TYY609 pKa = 10.93FGSGTANFYY618 pKa = 11.07LSDD621 pKa = 3.29ATYY624 pKa = 9.58TGDD627 pKa = 3.23VHH629 pKa = 7.81FEE631 pKa = 4.04SEE633 pKa = 4.25GFNFQSTQGQYY644 pKa = 10.53FGNLNFYY651 pKa = 9.93NNSGSAVFQNNSYY664 pKa = 10.76FSGSLLDD671 pKa = 4.27SDD673 pKa = 5.41NIDD676 pKa = 5.0LIIDD680 pKa = 4.03TSTIYY685 pKa = 10.19ISQDD689 pKa = 2.91SPITLNSLNITGASTLGVIISNSEE713 pKa = 4.36HH714 pKa = 7.0IDD716 pKa = 3.42TPYY719 pKa = 9.4ITATNTAIVGDD730 pKa = 4.48SVTLSSQFTNFVGGDD745 pKa = 3.57FSRR748 pKa = 11.84TILSAPSLDD757 pKa = 3.33VDD759 pKa = 3.39IDD761 pKa = 4.29SISFDD766 pKa = 3.41TSGLSWLYY774 pKa = 9.11NAEE777 pKa = 4.3YY778 pKa = 10.45ILDD781 pKa = 3.78QEE783 pKa = 4.6ADD785 pKa = 3.76GNQSISLQFSQKK797 pKa = 8.82TSEE800 pKa = 4.47DD801 pKa = 3.59LGLDD805 pKa = 3.47AQQDD809 pKa = 3.5RR810 pKa = 11.84TFASFIEE817 pKa = 4.71MISNQQNAGSEE828 pKa = 4.39FVKK831 pKa = 9.76ITDD834 pKa = 3.21EE835 pKa = 4.43TTFLEE840 pKa = 4.89AYY842 pKa = 10.0EE843 pKa = 4.8SILPQHH849 pKa = 6.73SDD851 pKa = 2.79VALRR855 pKa = 11.84HH856 pKa = 6.02LDD858 pKa = 3.16AHH860 pKa = 6.23TNTLNSMVSEE870 pKa = 4.34RR871 pKa = 11.84MALLRR876 pKa = 11.84DD877 pKa = 3.69SSNTSNGFWLQEE889 pKa = 4.11LVSQTTVDD897 pKa = 2.98ASEE900 pKa = 4.15NVNAYY905 pKa = 9.77SGRR908 pKa = 11.84GFGFTAGVDD917 pKa = 3.5RR918 pKa = 11.84RR919 pKa = 11.84IGFIDD924 pKa = 3.56SMGLTFSINDD934 pKa = 3.49EE935 pKa = 4.29KK936 pKa = 11.76YY937 pKa = 7.67EE938 pKa = 4.26TTTSAFNEE946 pKa = 4.27TSSTNFALGFYY957 pKa = 9.33IAEE960 pKa = 4.5RR961 pKa = 11.84IGFADD966 pKa = 4.11LQMSAQIGQTNFKK979 pKa = 10.24SEE981 pKa = 4.16RR982 pKa = 11.84EE983 pKa = 4.17VNFEE987 pKa = 4.18GFNSDD992 pKa = 5.54IEE994 pKa = 4.58GDD996 pKa = 3.49WSGLTQAYY1004 pKa = 6.97STQISSPQKK1013 pKa = 10.32LGWLRR1018 pKa = 11.84VTPHH1022 pKa = 6.64IGASYY1027 pKa = 10.79ISLNQDD1033 pKa = 2.65AYY1035 pKa = 10.7EE1036 pKa = 4.12EE1037 pKa = 4.25TASNGFNLAFSEE1049 pKa = 4.53SEE1051 pKa = 4.43SNKK1054 pKa = 8.8LTGSAGVSLGVFWPSNSGRR1073 pKa = 11.84NSFDD1077 pKa = 3.22LSEE1080 pKa = 4.25NQQSNSSLNGWHH1092 pKa = 6.76AAVDD1096 pKa = 3.66LGIRR1100 pKa = 11.84DD1101 pKa = 4.1TLSSTNYY1108 pKa = 9.74DD1109 pKa = 2.87AVANFVGYY1117 pKa = 10.73DD1118 pKa = 3.36SAFNVYY1124 pKa = 10.16SDD1126 pKa = 4.83EE1127 pKa = 4.48IFGQAFSTGISLVGIGDD1144 pKa = 4.06FASFRR1149 pKa = 11.84LSANAEE1155 pKa = 4.04LSEE1158 pKa = 4.19GASIYY1163 pKa = 10.86SGAASLRR1170 pKa = 11.84FKK1172 pKa = 10.96FF1173 pKa = 4.26
Molecular weight: 124.6 kDa
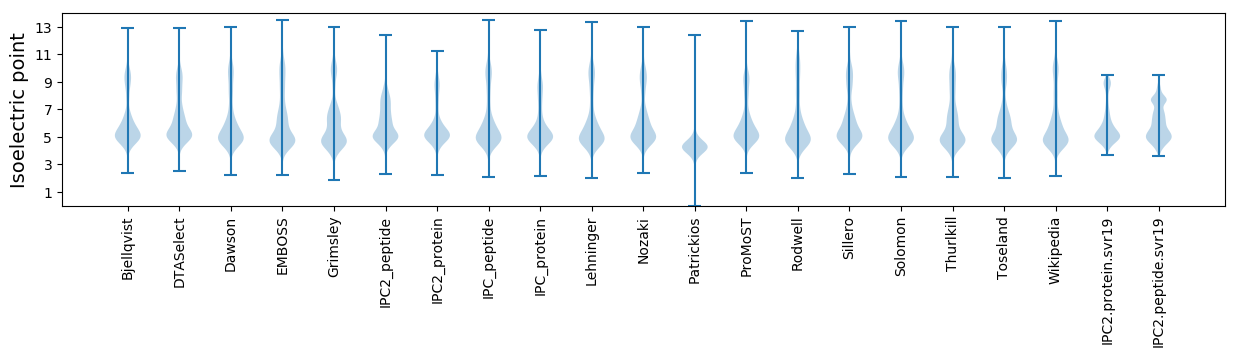
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6XQ63|C6XQ63_HIRBI Histidine kinase OS=Hirschia baltica (strain ATCC 49814 / DSM 5838 / IFAM 1418) OX=582402 GN=Hbal_0886 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATANGQKK29 pKa = 9.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84KK40 pKa = 8.79KK41 pKa = 10.63LSAA44 pKa = 3.95
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.62GFRR19 pKa = 11.84SRR21 pKa = 11.84MATANGQKK29 pKa = 9.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84KK40 pKa = 8.79KK41 pKa = 10.63LSAA44 pKa = 3.95
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1051554 |
30 |
1641 |
330.0 |
36.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.146 ± 0.055 | 0.884 ± 0.016 |
6.076 ± 0.035 | 6.558 ± 0.041 |
4.087 ± 0.032 | 7.596 ± 0.043 |
1.909 ± 0.023 | 6.253 ± 0.032 |
4.909 ± 0.037 | 9.433 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.579 ± 0.02 | 3.853 ± 0.028 |
4.424 ± 0.029 | 3.439 ± 0.024 |
5.203 ± 0.03 | 6.685 ± 0.037 |
5.398 ± 0.031 | 6.732 ± 0.034 |
1.281 ± 0.017 | 2.555 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
