
Lottia gigantea (Giant owl limpet)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Mollusca; Gastropoda; Patellogastropoda; Lottioidea; Lottiidae; Lottia
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
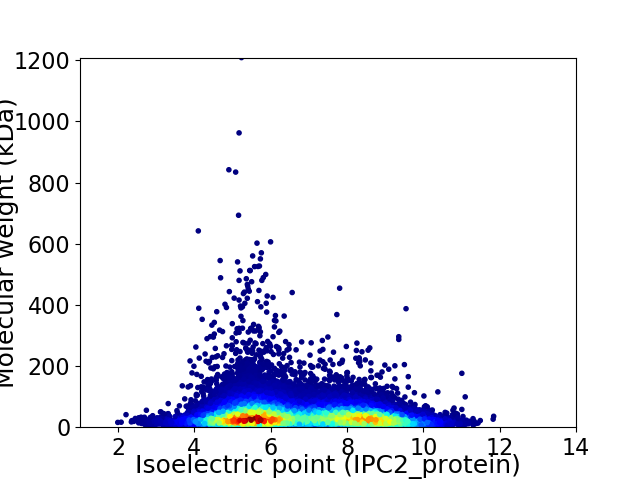
Virtual 2D-PAGE plot for 23675 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V3ZQS5|V3ZQS5_LOTGI Uncharacterized protein OS=Lottia gigantea OX=225164 GN=LOTGIDRAFT_235302 PE=3 SV=1
EEE2 pKa = 4.92DDD4 pKa = 3.65KK5 pKa = 11.27LLTRR9 pKa = 11.84VVIFEEE15 pKa = 4.49VSTLKKK21 pKa = 10.66KKK23 pKa = 10.92SIPDDD28 pKa = 3.63DD29 pKa = 3.59LLVLVQFVLLDDD41 pKa = 3.59GGSLCSSILSDDD53 pKa = 5.44DD54 pKa = 3.91VLDDD58 pKa = 3.86GGSLCSSILSDDD70 pKa = 5.44DD71 pKa = 3.91VLDDD75 pKa = 3.86GGSLCSSILSDDD87 pKa = 5.44DD88 pKa = 3.91VLDDD92 pKa = 3.86GGSLCSSILSDDD104 pKa = 5.44DD105 pKa = 3.91VLDDD109 pKa = 3.86GGSLCSSLLSDDD121 pKa = 5.46DD122 pKa = 4.36VLDDD126 pKa = 3.86GGSLCSSILSDDD138 pKa = 4.55DD139 pKa = 3.48GMTYYY144 pKa = 10.11SPPQSLLSYYY154 pKa = 10.48AAEEE158 pKa = 5.37MRR160 pKa = 11.84QHHH163 pKa = 6.88NDDD166 pKa = 4.66IEEE169 pKa = 4.89ISDDD173 pKa = 3.33HHH175 pKa = 6.19LTKKK179 pKa = 10.72K
EEE2 pKa = 4.92DDD4 pKa = 3.65KK5 pKa = 11.27LLTRR9 pKa = 11.84VVIFEEE15 pKa = 4.49VSTLKKK21 pKa = 10.66KKK23 pKa = 10.92SIPDDD28 pKa = 3.63DD29 pKa = 3.59LLVLVQFVLLDDD41 pKa = 3.59GGSLCSSILSDDD53 pKa = 5.44DD54 pKa = 3.91VLDDD58 pKa = 3.86GGSLCSSILSDDD70 pKa = 5.44DD71 pKa = 3.91VLDDD75 pKa = 3.86GGSLCSSILSDDD87 pKa = 5.44DD88 pKa = 3.91VLDDD92 pKa = 3.86GGSLCSSILSDDD104 pKa = 5.44DD105 pKa = 3.91VLDDD109 pKa = 3.86GGSLCSSLLSDDD121 pKa = 5.46DD122 pKa = 4.36VLDDD126 pKa = 3.86GGSLCSSILSDDD138 pKa = 4.55DD139 pKa = 3.48GMTYYY144 pKa = 10.11SPPQSLLSYYY154 pKa = 10.48AAEEE158 pKa = 5.37MRR160 pKa = 11.84QHHH163 pKa = 6.88NDDD166 pKa = 4.66IEEE169 pKa = 4.89ISDDD173 pKa = 3.33HHH175 pKa = 6.19LTKKK179 pKa = 10.72K
Molecular weight: 18.95 kDa
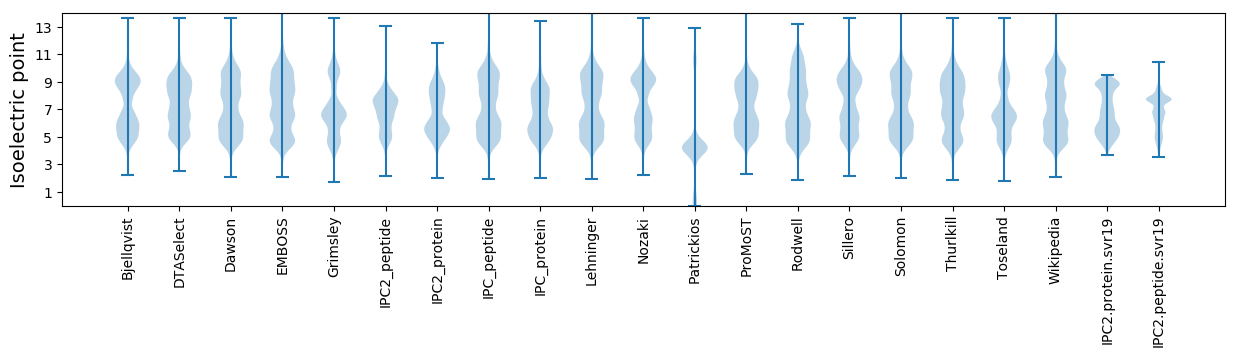
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V3ZXA6|V3ZXA6_LOTGI DNA replication licensing factor MCM7 OS=Lottia gigantea OX=225164 GN=MCM7 PE=3 SV=1
RR1 pKa = 7.42QNQFLRR7 pKa = 11.84INTTLFLNQFLQINITLFLNQFLQINITLFLNQFLRR43 pKa = 11.84INITLFLNQFLRR55 pKa = 11.84INITLFLNQFLRR67 pKa = 11.84INITLFLNLFLRR79 pKa = 11.84INITLFLNQFLRR91 pKa = 11.84INITLFLNQFLQINITLFLNQFLQINITLFLNQFLQINITLFLNQFLRR139 pKa = 11.84INIILFLNQFLQINITLFLNQFLQINITLFLNQFLRR175 pKa = 11.84INIILFLNQFLRR187 pKa = 11.84INITLFLNQFLRR199 pKa = 11.84INITLFLNLFLRR211 pKa = 11.84INITLFLNLFLRR223 pKa = 11.84INITLFF229 pKa = 3.39
RR1 pKa = 7.42QNQFLRR7 pKa = 11.84INTTLFLNQFLQINITLFLNQFLQINITLFLNQFLRR43 pKa = 11.84INITLFLNQFLRR55 pKa = 11.84INITLFLNQFLRR67 pKa = 11.84INITLFLNLFLRR79 pKa = 11.84INITLFLNQFLRR91 pKa = 11.84INITLFLNQFLQINITLFLNQFLQINITLFLNQFLQINITLFLNQFLRR139 pKa = 11.84INIILFLNQFLQINITLFLNQFLQINITLFLNQFLRR175 pKa = 11.84INIILFLNQFLRR187 pKa = 11.84INITLFLNQFLRR199 pKa = 11.84INITLFLNLFLRR211 pKa = 11.84INITLFLNLFLRR223 pKa = 11.84INITLFF229 pKa = 3.39
Molecular weight: 27.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8983955 |
49 |
10830 |
379.5 |
42.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.981 ± 0.014 | 2.317 ± 0.019 |
5.856 ± 0.016 | 6.233 ± 0.021 |
4.205 ± 0.013 | 5.515 ± 0.02 |
2.418 ± 0.01 | 6.454 ± 0.017 |
6.993 ± 0.02 | 8.83 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.315 ± 0.008 | 5.454 ± 0.016 |
4.546 ± 0.016 | 4.069 ± 0.015 |
4.761 ± 0.017 | 8.019 ± 0.027 |
6.074 ± 0.019 | 6.2 ± 0.014 |
1.107 ± 0.007 | 3.649 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
