
Bacillus sp. KQ-3
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; unclassified Bacillus (in: Bacteria)
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
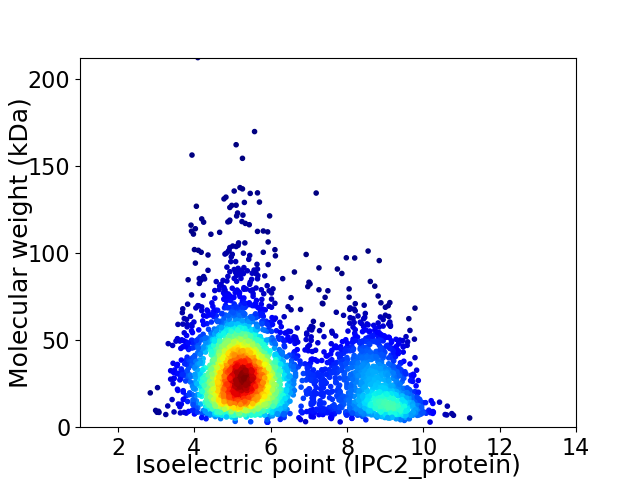
Virtual 2D-PAGE plot for 3944 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M7TN76|A0A3M7TN76_9BACI Aminoglycoside N(3)-acetyltransferase OS=Bacillus sp. KQ-3 OX=2483800 GN=EBO34_18005 PE=3 SV=1
MM1 pKa = 7.49TGLLFSTTAPAAQADD16 pKa = 4.02AEE18 pKa = 4.31DD19 pKa = 4.26TEE21 pKa = 4.75KK22 pKa = 11.02LVIMGTTDD30 pKa = 1.99IHH32 pKa = 8.52AYY34 pKa = 8.23MMPYY38 pKa = 10.43DD39 pKa = 4.15YY40 pKa = 11.13MNDD43 pKa = 3.85TVDD46 pKa = 3.55DD47 pKa = 4.43TYY49 pKa = 11.98GLTKK53 pKa = 10.41VHH55 pKa = 6.2TLVEE59 pKa = 4.86EE60 pKa = 4.12IRR62 pKa = 11.84SEE64 pKa = 4.35HH65 pKa = 6.71EE66 pKa = 3.77NTLLLDD72 pKa = 4.05NGDD75 pKa = 4.51IIQGSIFGDD84 pKa = 3.42MLALVDD90 pKa = 4.45PVGADD95 pKa = 3.67EE96 pKa = 4.5PHH98 pKa = 7.64VIMDD102 pKa = 4.29AMNMMGYY109 pKa = 10.23DD110 pKa = 3.4AATLGNHH117 pKa = 5.92EE118 pKa = 4.99FNFGLDD124 pKa = 3.74YY125 pKa = 11.17LDD127 pKa = 3.48EE128 pKa = 5.03TIAKK132 pKa = 10.13ADD134 pKa = 4.17FPWLSANVYY143 pKa = 10.87NDD145 pKa = 3.13GTEE148 pKa = 3.95DD149 pKa = 4.3LRR151 pKa = 11.84YY152 pKa = 10.22DD153 pKa = 3.95PYY155 pKa = 11.58AIIEE159 pKa = 4.17KK160 pKa = 9.93VINGEE165 pKa = 4.11TLNVGVIGLVPPQIMTWDD183 pKa = 3.63RR184 pKa = 11.84VHH186 pKa = 7.59LDD188 pKa = 2.98GNVYY192 pKa = 10.29VNEE195 pKa = 4.06IVEE198 pKa = 4.33SAKK201 pKa = 10.36TYY203 pKa = 9.04VPQMKK208 pKa = 10.32EE209 pKa = 3.46EE210 pKa = 4.24GADD213 pKa = 3.03IVVAVAHH220 pKa = 6.19SGINPAEE227 pKa = 4.44DD228 pKa = 3.31ASEE231 pKa = 3.89NAGYY235 pKa = 10.06QLSQVEE241 pKa = 5.3GIDD244 pKa = 4.29AMILGHH250 pKa = 5.39QHH252 pKa = 5.81NVFPGTDD259 pKa = 3.24AYY261 pKa = 11.46SGIDD265 pKa = 4.07GIDD268 pKa = 3.44VEE270 pKa = 4.82AGTVNGVPTVMPGAWGSHH288 pKa = 5.55LGVIEE293 pKa = 5.37LDD295 pKa = 3.57LTNTDD300 pKa = 3.73GQWEE304 pKa = 4.37VAGFDD309 pKa = 3.45SEE311 pKa = 4.44ARR313 pKa = 11.84EE314 pKa = 3.85TAAYY318 pKa = 7.98EE319 pKa = 3.79PHH321 pKa = 7.4PDD323 pKa = 3.13MVSLVEE329 pKa = 4.2EE330 pKa = 4.11THH332 pKa = 6.11EE333 pKa = 3.98ATIEE337 pKa = 4.19YY338 pKa = 10.19VNSPVGDD345 pKa = 3.6TATDD349 pKa = 3.88LNTFFSRR356 pKa = 11.84VMDD359 pKa = 3.83NEE361 pKa = 4.07VVQLVNDD368 pKa = 3.78AQLDD372 pKa = 3.82YY373 pKa = 10.76MEE375 pKa = 5.24QNFASTEE382 pKa = 4.06YY383 pKa = 10.57EE384 pKa = 4.06DD385 pKa = 3.82MAMLSAAAPFRR396 pKa = 11.84AGRR399 pKa = 11.84GGAYY403 pKa = 9.56TNVEE407 pKa = 4.27EE408 pKa = 4.42GSIAIRR414 pKa = 11.84DD415 pKa = 3.63VNDD418 pKa = 2.99IYY420 pKa = 11.46VYY422 pKa = 10.68PNTLHH427 pKa = 5.3VVKK430 pKa = 11.12VNGTEE435 pKa = 3.73LHH437 pKa = 5.83NWLEE441 pKa = 4.02RR442 pKa = 11.84SAEE445 pKa = 3.98NFLQIDD451 pKa = 4.08PDD453 pKa = 3.88SSDD456 pKa = 2.99RR457 pKa = 11.84QYY459 pKa = 11.84VINTDD464 pKa = 3.76FASFDD469 pKa = 3.51FDD471 pKa = 3.94TIEE474 pKa = 4.1GVEE477 pKa = 3.99YY478 pKa = 10.59QIDD481 pKa = 3.62VTQPVGEE488 pKa = 4.42RR489 pKa = 11.84IVGLTYY495 pKa = 10.38EE496 pKa = 4.67GEE498 pKa = 4.33DD499 pKa = 3.25VTEE502 pKa = 3.94EE503 pKa = 4.7DD504 pKa = 3.96EE505 pKa = 5.87FLVATNNYY513 pKa = 8.71RR514 pKa = 11.84AGGGGAHH521 pKa = 6.97IDD523 pKa = 3.64GVEE526 pKa = 4.16TVLASTDD533 pKa = 3.29EE534 pKa = 3.97NRR536 pKa = 11.84EE537 pKa = 3.9VIIDD541 pKa = 3.84YY542 pKa = 10.71IRR544 pKa = 11.84DD545 pKa = 3.38HH546 pKa = 7.38DD547 pKa = 4.4GPLEE551 pKa = 4.15VEE553 pKa = 4.55VTNNWSLVPFEE564 pKa = 4.51TDD566 pKa = 2.7AQVVFEE572 pKa = 5.09SDD574 pKa = 3.39LQGEE578 pKa = 4.5PLADD582 pKa = 3.4TTPFVSFVEE591 pKa = 4.69TVNDD595 pKa = 3.73TTGLFQIHH603 pKa = 5.86PQEE606 pKa = 4.25HH607 pKa = 6.24VEE609 pKa = 5.09DD610 pKa = 5.45GDD612 pKa = 6.18DD613 pKa = 3.6EE614 pKa = 6.82DD615 pKa = 5.51EE616 pKa = 5.42DD617 pKa = 4.47DD618 pKa = 4.9RR619 pKa = 11.84PGNRR623 pKa = 11.84PPHH626 pKa = 6.57AGQPGPPPHH635 pKa = 7.17AGQPGPPPHH644 pKa = 7.07ANN646 pKa = 3.0
MM1 pKa = 7.49TGLLFSTTAPAAQADD16 pKa = 4.02AEE18 pKa = 4.31DD19 pKa = 4.26TEE21 pKa = 4.75KK22 pKa = 11.02LVIMGTTDD30 pKa = 1.99IHH32 pKa = 8.52AYY34 pKa = 8.23MMPYY38 pKa = 10.43DD39 pKa = 4.15YY40 pKa = 11.13MNDD43 pKa = 3.85TVDD46 pKa = 3.55DD47 pKa = 4.43TYY49 pKa = 11.98GLTKK53 pKa = 10.41VHH55 pKa = 6.2TLVEE59 pKa = 4.86EE60 pKa = 4.12IRR62 pKa = 11.84SEE64 pKa = 4.35HH65 pKa = 6.71EE66 pKa = 3.77NTLLLDD72 pKa = 4.05NGDD75 pKa = 4.51IIQGSIFGDD84 pKa = 3.42MLALVDD90 pKa = 4.45PVGADD95 pKa = 3.67EE96 pKa = 4.5PHH98 pKa = 7.64VIMDD102 pKa = 4.29AMNMMGYY109 pKa = 10.23DD110 pKa = 3.4AATLGNHH117 pKa = 5.92EE118 pKa = 4.99FNFGLDD124 pKa = 3.74YY125 pKa = 11.17LDD127 pKa = 3.48EE128 pKa = 5.03TIAKK132 pKa = 10.13ADD134 pKa = 4.17FPWLSANVYY143 pKa = 10.87NDD145 pKa = 3.13GTEE148 pKa = 3.95DD149 pKa = 4.3LRR151 pKa = 11.84YY152 pKa = 10.22DD153 pKa = 3.95PYY155 pKa = 11.58AIIEE159 pKa = 4.17KK160 pKa = 9.93VINGEE165 pKa = 4.11TLNVGVIGLVPPQIMTWDD183 pKa = 3.63RR184 pKa = 11.84VHH186 pKa = 7.59LDD188 pKa = 2.98GNVYY192 pKa = 10.29VNEE195 pKa = 4.06IVEE198 pKa = 4.33SAKK201 pKa = 10.36TYY203 pKa = 9.04VPQMKK208 pKa = 10.32EE209 pKa = 3.46EE210 pKa = 4.24GADD213 pKa = 3.03IVVAVAHH220 pKa = 6.19SGINPAEE227 pKa = 4.44DD228 pKa = 3.31ASEE231 pKa = 3.89NAGYY235 pKa = 10.06QLSQVEE241 pKa = 5.3GIDD244 pKa = 4.29AMILGHH250 pKa = 5.39QHH252 pKa = 5.81NVFPGTDD259 pKa = 3.24AYY261 pKa = 11.46SGIDD265 pKa = 4.07GIDD268 pKa = 3.44VEE270 pKa = 4.82AGTVNGVPTVMPGAWGSHH288 pKa = 5.55LGVIEE293 pKa = 5.37LDD295 pKa = 3.57LTNTDD300 pKa = 3.73GQWEE304 pKa = 4.37VAGFDD309 pKa = 3.45SEE311 pKa = 4.44ARR313 pKa = 11.84EE314 pKa = 3.85TAAYY318 pKa = 7.98EE319 pKa = 3.79PHH321 pKa = 7.4PDD323 pKa = 3.13MVSLVEE329 pKa = 4.2EE330 pKa = 4.11THH332 pKa = 6.11EE333 pKa = 3.98ATIEE337 pKa = 4.19YY338 pKa = 10.19VNSPVGDD345 pKa = 3.6TATDD349 pKa = 3.88LNTFFSRR356 pKa = 11.84VMDD359 pKa = 3.83NEE361 pKa = 4.07VVQLVNDD368 pKa = 3.78AQLDD372 pKa = 3.82YY373 pKa = 10.76MEE375 pKa = 5.24QNFASTEE382 pKa = 4.06YY383 pKa = 10.57EE384 pKa = 4.06DD385 pKa = 3.82MAMLSAAAPFRR396 pKa = 11.84AGRR399 pKa = 11.84GGAYY403 pKa = 9.56TNVEE407 pKa = 4.27EE408 pKa = 4.42GSIAIRR414 pKa = 11.84DD415 pKa = 3.63VNDD418 pKa = 2.99IYY420 pKa = 11.46VYY422 pKa = 10.68PNTLHH427 pKa = 5.3VVKK430 pKa = 11.12VNGTEE435 pKa = 3.73LHH437 pKa = 5.83NWLEE441 pKa = 4.02RR442 pKa = 11.84SAEE445 pKa = 3.98NFLQIDD451 pKa = 4.08PDD453 pKa = 3.88SSDD456 pKa = 2.99RR457 pKa = 11.84QYY459 pKa = 11.84VINTDD464 pKa = 3.76FASFDD469 pKa = 3.51FDD471 pKa = 3.94TIEE474 pKa = 4.1GVEE477 pKa = 3.99YY478 pKa = 10.59QIDD481 pKa = 3.62VTQPVGEE488 pKa = 4.42RR489 pKa = 11.84IVGLTYY495 pKa = 10.38EE496 pKa = 4.67GEE498 pKa = 4.33DD499 pKa = 3.25VTEE502 pKa = 3.94EE503 pKa = 4.7DD504 pKa = 3.96EE505 pKa = 5.87FLVATNNYY513 pKa = 8.71RR514 pKa = 11.84AGGGGAHH521 pKa = 6.97IDD523 pKa = 3.64GVEE526 pKa = 4.16TVLASTDD533 pKa = 3.29EE534 pKa = 3.97NRR536 pKa = 11.84EE537 pKa = 3.9VIIDD541 pKa = 3.84YY542 pKa = 10.71IRR544 pKa = 11.84DD545 pKa = 3.38HH546 pKa = 7.38DD547 pKa = 4.4GPLEE551 pKa = 4.15VEE553 pKa = 4.55VTNNWSLVPFEE564 pKa = 4.51TDD566 pKa = 2.7AQVVFEE572 pKa = 5.09SDD574 pKa = 3.39LQGEE578 pKa = 4.5PLADD582 pKa = 3.4TTPFVSFVEE591 pKa = 4.69TVNDD595 pKa = 3.73TTGLFQIHH603 pKa = 5.86PQEE606 pKa = 4.25HH607 pKa = 6.24VEE609 pKa = 5.09DD610 pKa = 5.45GDD612 pKa = 6.18DD613 pKa = 3.6EE614 pKa = 6.82DD615 pKa = 5.51EE616 pKa = 5.42DD617 pKa = 4.47DD618 pKa = 4.9RR619 pKa = 11.84PGNRR623 pKa = 11.84PPHH626 pKa = 6.57AGQPGPPPHH635 pKa = 7.17AGQPGPPPHH644 pKa = 7.07ANN646 pKa = 3.0
Molecular weight: 70.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M7TQW7|A0A3M7TQW7_9BACI Flagellar protein FlgN OS=Bacillus sp. KQ-3 OX=2483800 GN=EBO34_12640 PE=4 SV=1
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.18RR13 pKa = 11.84KK14 pKa = 8.6KK15 pKa = 9.25VHH17 pKa = 5.37GFRR20 pKa = 11.84NRR22 pKa = 11.84MSTKK26 pKa = 9.22NGRR29 pKa = 11.84RR30 pKa = 11.84VLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.69GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.18RR13 pKa = 11.84KK14 pKa = 8.6KK15 pKa = 9.25VHH17 pKa = 5.37GFRR20 pKa = 11.84NRR22 pKa = 11.84MSTKK26 pKa = 9.22NGRR29 pKa = 11.84RR30 pKa = 11.84VLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.69GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1152186 |
25 |
1895 |
292.1 |
32.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.264 ± 0.043 | 0.698 ± 0.012 |
5.532 ± 0.036 | 8.08 ± 0.056 |
4.527 ± 0.032 | 7.365 ± 0.032 |
2.187 ± 0.02 | 6.796 ± 0.041 |
6.151 ± 0.046 | 9.514 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.887 ± 0.02 | 3.923 ± 0.026 |
3.769 ± 0.023 | 3.362 ± 0.024 |
4.589 ± 0.029 | 5.919 ± 0.029 |
5.516 ± 0.023 | 7.456 ± 0.03 |
1.088 ± 0.014 | 3.377 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
