
Flavobacterium sp. 316
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; unclassified Flavobacterium
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
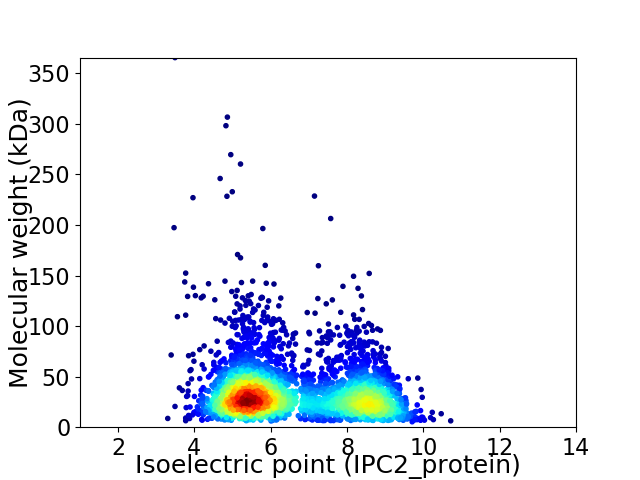
Virtual 2D-PAGE plot for 3356 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
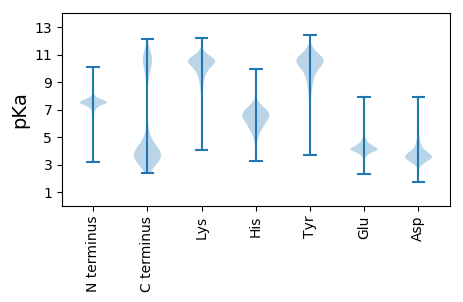
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D6TLC6|A0A0D6TLC6_9FLAO Uncharacterized protein OS=Flavobacterium sp. 316 OX=1603293 GN=SY27_09405 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 10.25SLKK5 pKa = 10.28INLLKK10 pKa = 10.52KK11 pKa = 9.45IQYY14 pKa = 10.74AFIAFLYY21 pKa = 10.42CIYY24 pKa = 10.06CHH26 pKa = 6.57NYY28 pKa = 9.77EE29 pKa = 4.94ILAQNQLYY37 pKa = 7.23PTTFDD42 pKa = 4.13LSTLNGSNGFEE53 pKa = 4.05VSGLSTNLQFGAEE66 pKa = 4.15SQFIGDD72 pKa = 3.95INNDD76 pKa = 3.42GLEE79 pKa = 4.62DD80 pKa = 3.34IAFGSGTATASGLQYY95 pKa = 11.18AGAAYY100 pKa = 9.51IIFGSSVPYY109 pKa = 9.58PSPFDD114 pKa = 4.53LSTLDD119 pKa = 3.43GSNGFTVDD127 pKa = 4.12GIAYY131 pKa = 9.39DD132 pKa = 3.86EE133 pKa = 4.42RR134 pKa = 11.84RR135 pKa = 11.84GATITGPGDD144 pKa = 3.38INGDD148 pKa = 3.95GIDD151 pKa = 4.16DD152 pKa = 5.4LIIGSSSTSPDD163 pKa = 3.19EE164 pKa = 3.63IVIYY168 pKa = 8.96GTTSFPALITINDD181 pKa = 3.42INGTNGFLIDD191 pKa = 4.16TPGSNQITGLGDD203 pKa = 3.39VNGDD207 pKa = 4.17GINDD211 pKa = 4.54FIIGTPHH218 pKa = 7.15WSGQAWIIFGRR229 pKa = 11.84SSNFPTSIDD238 pKa = 3.69ASWLDD243 pKa = 3.38GTKK246 pKa = 10.63GFKK249 pKa = 9.79TSSFPGSRR257 pKa = 11.84PSYY260 pKa = 9.8KK261 pKa = 10.45VGGAGDD267 pKa = 3.7INNDD271 pKa = 3.03GFKK274 pKa = 10.72DD275 pKa = 3.43ILIGNWDD282 pKa = 3.75STNPAISYY290 pKa = 10.25LLFGKK295 pKa = 7.62GTPFSPLVDD304 pKa = 3.87LTTLDD309 pKa = 3.65GTDD312 pKa = 3.99GFSIDD317 pKa = 3.75NNGGNFLTFVGTLGDD332 pKa = 3.92INNDD336 pKa = 3.88GIDD339 pKa = 3.79DD340 pKa = 4.6FFSEE344 pKa = 4.4TNAIYY349 pKa = 10.44GSTDD353 pKa = 3.11PFPAHH358 pKa = 6.56FPLSSLNGTDD368 pKa = 4.46GFTLPGQLTSAGIGDD383 pKa = 3.91INMDD387 pKa = 4.5GIDD390 pKa = 4.61DD391 pKa = 5.25FISVYY396 pKa = 10.41GSSGRR401 pKa = 11.84AYY403 pKa = 9.47IVFGSTSGFPNPINEE418 pKa = 4.23TTLNGVNGFIIEE430 pKa = 4.35GLNPTNIARR439 pKa = 11.84PVSGNGDD446 pKa = 3.31INGDD450 pKa = 3.96GISDD454 pKa = 3.77FVVGSPYY461 pKa = 10.69TPTTQIGKK469 pKa = 9.72SYY471 pKa = 10.83VIFGGDD477 pKa = 3.53HH478 pKa = 5.87YY479 pKa = 11.92AMPLNIEE486 pKa = 4.19YY487 pKa = 8.97PQTEE491 pKa = 4.51NVTPTALTLIVNGPEE506 pKa = 3.81TGTIHH511 pKa = 6.64YY512 pKa = 10.51AIFPGNNAGTVTHH525 pKa = 6.83NDD527 pKa = 2.91ILNGTGAIANGNFLMNVQNGNVNNIVSSLPTNSSFDD563 pKa = 3.34VYY565 pKa = 11.42LFLEE569 pKa = 4.81DD570 pKa = 4.83NAGNWTQIYY579 pKa = 10.28DD580 pKa = 4.04LNNVLDD586 pKa = 4.27NEE588 pKa = 4.26DD589 pKa = 4.81FILSEE594 pKa = 3.96TDD596 pKa = 2.82IYY598 pKa = 10.8PNPTTSSINVNLTTYY613 pKa = 8.96ATYY616 pKa = 10.95EE617 pKa = 4.39LITINGQVIQKK628 pKa = 10.8GEE630 pKa = 4.4LINGKK635 pKa = 9.13NVVDD639 pKa = 4.04MSSLSKK645 pKa = 10.68GLYY648 pKa = 9.19FLYY651 pKa = 10.87VKK653 pKa = 9.73TDD655 pKa = 2.73AGTATKK661 pKa = 10.53KK662 pKa = 10.29IMKK665 pKa = 9.71KK666 pKa = 10.39
MM1 pKa = 7.64KK2 pKa = 10.25SLKK5 pKa = 10.28INLLKK10 pKa = 10.52KK11 pKa = 9.45IQYY14 pKa = 10.74AFIAFLYY21 pKa = 10.42CIYY24 pKa = 10.06CHH26 pKa = 6.57NYY28 pKa = 9.77EE29 pKa = 4.94ILAQNQLYY37 pKa = 7.23PTTFDD42 pKa = 4.13LSTLNGSNGFEE53 pKa = 4.05VSGLSTNLQFGAEE66 pKa = 4.15SQFIGDD72 pKa = 3.95INNDD76 pKa = 3.42GLEE79 pKa = 4.62DD80 pKa = 3.34IAFGSGTATASGLQYY95 pKa = 11.18AGAAYY100 pKa = 9.51IIFGSSVPYY109 pKa = 9.58PSPFDD114 pKa = 4.53LSTLDD119 pKa = 3.43GSNGFTVDD127 pKa = 4.12GIAYY131 pKa = 9.39DD132 pKa = 3.86EE133 pKa = 4.42RR134 pKa = 11.84RR135 pKa = 11.84GATITGPGDD144 pKa = 3.38INGDD148 pKa = 3.95GIDD151 pKa = 4.16DD152 pKa = 5.4LIIGSSSTSPDD163 pKa = 3.19EE164 pKa = 3.63IVIYY168 pKa = 8.96GTTSFPALITINDD181 pKa = 3.42INGTNGFLIDD191 pKa = 4.16TPGSNQITGLGDD203 pKa = 3.39VNGDD207 pKa = 4.17GINDD211 pKa = 4.54FIIGTPHH218 pKa = 7.15WSGQAWIIFGRR229 pKa = 11.84SSNFPTSIDD238 pKa = 3.69ASWLDD243 pKa = 3.38GTKK246 pKa = 10.63GFKK249 pKa = 9.79TSSFPGSRR257 pKa = 11.84PSYY260 pKa = 9.8KK261 pKa = 10.45VGGAGDD267 pKa = 3.7INNDD271 pKa = 3.03GFKK274 pKa = 10.72DD275 pKa = 3.43ILIGNWDD282 pKa = 3.75STNPAISYY290 pKa = 10.25LLFGKK295 pKa = 7.62GTPFSPLVDD304 pKa = 3.87LTTLDD309 pKa = 3.65GTDD312 pKa = 3.99GFSIDD317 pKa = 3.75NNGGNFLTFVGTLGDD332 pKa = 3.92INNDD336 pKa = 3.88GIDD339 pKa = 3.79DD340 pKa = 4.6FFSEE344 pKa = 4.4TNAIYY349 pKa = 10.44GSTDD353 pKa = 3.11PFPAHH358 pKa = 6.56FPLSSLNGTDD368 pKa = 4.46GFTLPGQLTSAGIGDD383 pKa = 3.91INMDD387 pKa = 4.5GIDD390 pKa = 4.61DD391 pKa = 5.25FISVYY396 pKa = 10.41GSSGRR401 pKa = 11.84AYY403 pKa = 9.47IVFGSTSGFPNPINEE418 pKa = 4.23TTLNGVNGFIIEE430 pKa = 4.35GLNPTNIARR439 pKa = 11.84PVSGNGDD446 pKa = 3.31INGDD450 pKa = 3.96GISDD454 pKa = 3.77FVVGSPYY461 pKa = 10.69TPTTQIGKK469 pKa = 9.72SYY471 pKa = 10.83VIFGGDD477 pKa = 3.53HH478 pKa = 5.87YY479 pKa = 11.92AMPLNIEE486 pKa = 4.19YY487 pKa = 8.97PQTEE491 pKa = 4.51NVTPTALTLIVNGPEE506 pKa = 3.81TGTIHH511 pKa = 6.64YY512 pKa = 10.51AIFPGNNAGTVTHH525 pKa = 6.83NDD527 pKa = 2.91ILNGTGAIANGNFLMNVQNGNVNNIVSSLPTNSSFDD563 pKa = 3.34VYY565 pKa = 11.42LFLEE569 pKa = 4.81DD570 pKa = 4.83NAGNWTQIYY579 pKa = 10.28DD580 pKa = 4.04LNNVLDD586 pKa = 4.27NEE588 pKa = 4.26DD589 pKa = 4.81FILSEE594 pKa = 3.96TDD596 pKa = 2.82IYY598 pKa = 10.8PNPTTSSINVNLTTYY613 pKa = 8.96ATYY616 pKa = 10.95EE617 pKa = 4.39LITINGQVIQKK628 pKa = 10.8GEE630 pKa = 4.4LINGKK635 pKa = 9.13NVVDD639 pKa = 4.04MSSLSKK645 pKa = 10.68GLYY648 pKa = 9.19FLYY651 pKa = 10.87VKK653 pKa = 9.73TDD655 pKa = 2.73AGTATKK661 pKa = 10.53KK662 pKa = 10.29IMKK665 pKa = 9.71KK666 pKa = 10.39
Molecular weight: 70.7 kDa
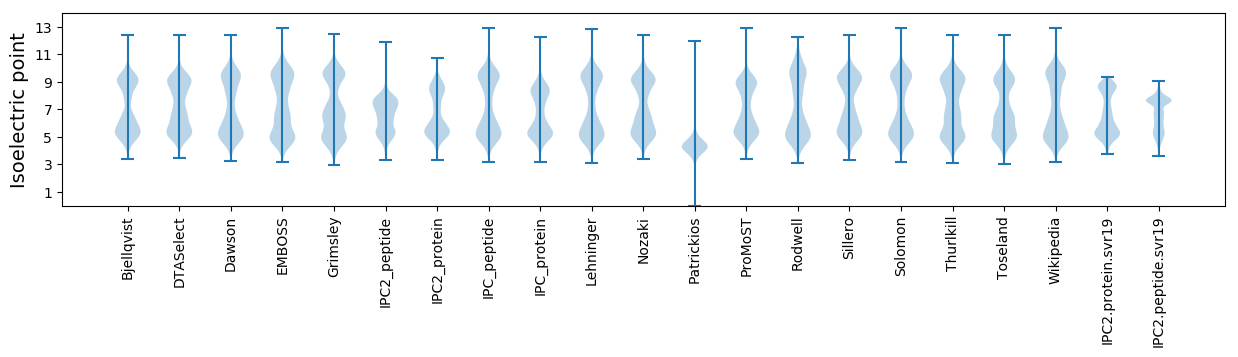
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D6TPJ8|A0A0D6TPJ8_9FLAO Pseudouridylate synthase OS=Flavobacterium sp. 316 OX=1603293 GN=SY27_08435 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.91HH17 pKa = 4.39GFMDD21 pKa = 4.5RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.41KK42 pKa = 10.14LTVSSEE48 pKa = 3.92PRR50 pKa = 11.84HH51 pKa = 5.77KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1094763 |
46 |
3465 |
326.2 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.482 ± 0.048 | 0.787 ± 0.013 |
5.266 ± 0.034 | 6.895 ± 0.046 |
5.493 ± 0.037 | 5.838 ± 0.047 |
1.636 ± 0.018 | 8.916 ± 0.043 |
8.55 ± 0.06 | 9.142 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.046 ± 0.022 | 7.067 ± 0.048 |
3.142 ± 0.025 | 3.318 ± 0.025 |
2.987 ± 0.027 | 6.568 ± 0.038 |
5.829 ± 0.045 | 5.737 ± 0.037 |
0.98 ± 0.015 | 4.316 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
