
La Piedad-Michoacan-Mexico virus (LPMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Orthorubulavirus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
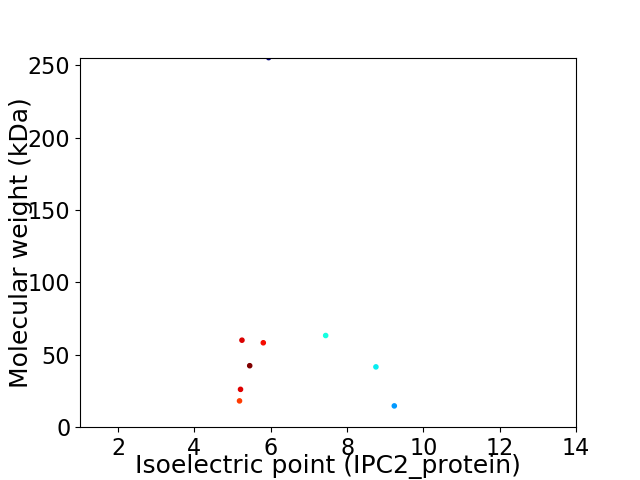
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
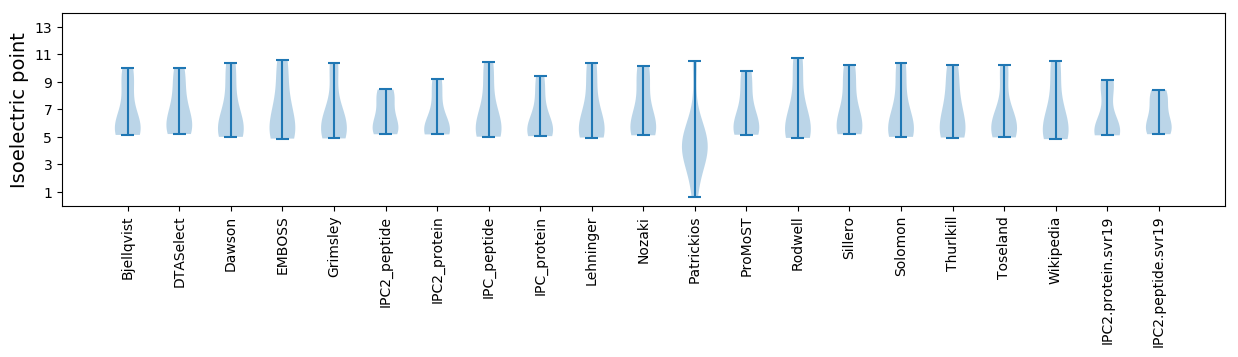
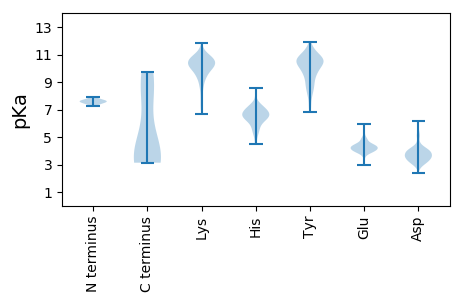
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q917N9|Q917N9_LPMV Isoform of A7E3E6 Non-structural protein V OS=La Piedad-Michoacan-Mexico virus OX=2560645 GN=P PE=2 SV=1
MM1 pKa = 7.66ASSSLSFSDD10 pKa = 5.1GEE12 pKa = 4.06ITEE15 pKa = 4.31LLEE18 pKa = 4.49TGLGTIEE25 pKa = 4.28SIEE28 pKa = 4.23RR29 pKa = 11.84MVAAKK34 pKa = 10.08GGPDD38 pKa = 3.34GGIDD42 pKa = 3.71PEE44 pKa = 4.42SQPGQRR50 pKa = 11.84GLPTPTPRR58 pKa = 11.84TTSTPTAAGSASATLEE74 pKa = 3.85LSPEE78 pKa = 3.82GGAIKK83 pKa = 10.15KK84 pKa = 10.24APRR87 pKa = 11.84AHH89 pKa = 6.38PTLPNPLGQEE99 pKa = 3.99EE100 pKa = 4.85RR101 pKa = 11.84PGNPLSTFTPVRR113 pKa = 11.84GSSSTHH119 pKa = 6.64DD120 pKa = 4.3PLPGSRR126 pKa = 11.84PGSEE130 pKa = 4.33VYY132 pKa = 10.54EE133 pKa = 4.62GDD135 pKa = 3.06LMARR139 pKa = 11.84ARR141 pKa = 11.84SEE143 pKa = 4.39LVTRR147 pKa = 11.84WSDD150 pKa = 3.51EE151 pKa = 3.99EE152 pKa = 4.53GDD154 pKa = 3.95PVPTRR159 pKa = 11.84VLQSTFKK166 pKa = 10.66RR167 pKa = 11.84GADD170 pKa = 2.8RR171 pKa = 11.84KK172 pKa = 9.75GANPGEE178 pKa = 4.35PGHH181 pKa = 6.15RR182 pKa = 11.84RR183 pKa = 11.84EE184 pKa = 4.3YY185 pKa = 10.72SIGWVCGTVRR195 pKa = 11.84VLEE198 pKa = 4.31WCNPACSPISMEE210 pKa = 3.56PRR212 pKa = 11.84YY213 pKa = 9.01YY214 pKa = 10.51QCTCGTCPAKK224 pKa = 10.6CPQCAGDD231 pKa = 3.73NGIVEE236 pKa = 4.94SNRR239 pKa = 11.84GPHH242 pKa = 6.94DD243 pKa = 3.62GAGNEE248 pKa = 4.29SS249 pKa = 3.43
MM1 pKa = 7.66ASSSLSFSDD10 pKa = 5.1GEE12 pKa = 4.06ITEE15 pKa = 4.31LLEE18 pKa = 4.49TGLGTIEE25 pKa = 4.28SIEE28 pKa = 4.23RR29 pKa = 11.84MVAAKK34 pKa = 10.08GGPDD38 pKa = 3.34GGIDD42 pKa = 3.71PEE44 pKa = 4.42SQPGQRR50 pKa = 11.84GLPTPTPRR58 pKa = 11.84TTSTPTAAGSASATLEE74 pKa = 3.85LSPEE78 pKa = 3.82GGAIKK83 pKa = 10.15KK84 pKa = 10.24APRR87 pKa = 11.84AHH89 pKa = 6.38PTLPNPLGQEE99 pKa = 3.99EE100 pKa = 4.85RR101 pKa = 11.84PGNPLSTFTPVRR113 pKa = 11.84GSSSTHH119 pKa = 6.64DD120 pKa = 4.3PLPGSRR126 pKa = 11.84PGSEE130 pKa = 4.33VYY132 pKa = 10.54EE133 pKa = 4.62GDD135 pKa = 3.06LMARR139 pKa = 11.84ARR141 pKa = 11.84SEE143 pKa = 4.39LVTRR147 pKa = 11.84WSDD150 pKa = 3.51EE151 pKa = 3.99EE152 pKa = 4.53GDD154 pKa = 3.95PVPTRR159 pKa = 11.84VLQSTFKK166 pKa = 10.66RR167 pKa = 11.84GADD170 pKa = 2.8RR171 pKa = 11.84KK172 pKa = 9.75GANPGEE178 pKa = 4.35PGHH181 pKa = 6.15RR182 pKa = 11.84RR183 pKa = 11.84EE184 pKa = 4.3YY185 pKa = 10.72SIGWVCGTVRR195 pKa = 11.84VLEE198 pKa = 4.31WCNPACSPISMEE210 pKa = 3.56PRR212 pKa = 11.84YY213 pKa = 9.01YY214 pKa = 10.51QCTCGTCPAKK224 pKa = 10.6CPQCAGDD231 pKa = 3.73NGIVEE236 pKa = 4.94SNRR239 pKa = 11.84GPHH242 pKa = 6.94DD243 pKa = 3.62GAGNEE248 pKa = 4.29SS249 pKa = 3.43
Molecular weight: 26.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q917N9|Q917N9_LPMV Isoform of A7E3E6 Non-structural protein V OS=La Piedad-Michoacan-Mexico virus OX=2560645 GN=P PE=2 SV=1
MM1 pKa = 7.48EE2 pKa = 5.94RR3 pKa = 11.84LLNYY7 pKa = 8.15WKK9 pKa = 10.18QDD11 pKa = 2.96WALLRR16 pKa = 11.84VSRR19 pKa = 11.84GWWRR23 pKa = 11.84QRR25 pKa = 11.84VGLTGEE31 pKa = 4.68LTQSPNLDD39 pKa = 3.13RR40 pKa = 11.84GDD42 pKa = 3.69FRR44 pKa = 11.84LPPPGQHH51 pKa = 5.01QHH53 pKa = 6.75PPLQEE58 pKa = 3.91VPLLLLSLVRR68 pKa = 11.84KK69 pKa = 9.39EE70 pKa = 4.29EE71 pKa = 3.69QSKK74 pKa = 9.44RR75 pKa = 11.84HH76 pKa = 6.01LGRR79 pKa = 11.84TLHH82 pKa = 6.65SPTPLGKK89 pKa = 10.32KK90 pKa = 9.4NALEE94 pKa = 4.09TPYY97 pKa = 10.55PHH99 pKa = 7.2LPQSEE104 pKa = 4.47VHH106 pKa = 6.45HH107 pKa = 6.65PLMIPSLEE115 pKa = 4.1VVLDD119 pKa = 3.71QKK121 pKa = 10.84FMRR124 pKa = 11.84AII126 pKa = 3.65
MM1 pKa = 7.48EE2 pKa = 5.94RR3 pKa = 11.84LLNYY7 pKa = 8.15WKK9 pKa = 10.18QDD11 pKa = 2.96WALLRR16 pKa = 11.84VSRR19 pKa = 11.84GWWRR23 pKa = 11.84QRR25 pKa = 11.84VGLTGEE31 pKa = 4.68LTQSPNLDD39 pKa = 3.13RR40 pKa = 11.84GDD42 pKa = 3.69FRR44 pKa = 11.84LPPPGQHH51 pKa = 5.01QHH53 pKa = 6.75PPLQEE58 pKa = 3.91VPLLLLSLVRR68 pKa = 11.84KK69 pKa = 9.39EE70 pKa = 4.29EE71 pKa = 3.69QSKK74 pKa = 9.44RR75 pKa = 11.84HH76 pKa = 6.01LGRR79 pKa = 11.84TLHH82 pKa = 6.65SPTPLGKK89 pKa = 10.32KK90 pKa = 9.4NALEE94 pKa = 4.09TPYY97 pKa = 10.55PHH99 pKa = 7.2LPQSEE104 pKa = 4.47VHH106 pKa = 6.45HH107 pKa = 6.65PLMIPSLEE115 pKa = 4.1VVLDD119 pKa = 3.71QKK121 pKa = 10.84FMRR124 pKa = 11.84AII126 pKa = 3.65
Molecular weight: 14.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5235 |
126 |
2251 |
581.7 |
64.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.813 ± 0.976 | 1.796 ± 0.307 |
4.394 ± 0.462 | 5.444 ± 0.54 |
3.362 ± 0.481 | 6.399 ± 0.933 |
2.006 ± 0.349 | 6.38 ± 0.543 |
3.572 ± 0.527 | 10.869 ± 0.914 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.541 ± 0.178 | 3.859 ± 0.352 |
6.418 ± 1.192 | 4.852 ± 0.299 |
5.788 ± 0.442 | 7.335 ± 0.77 |
6.972 ± 0.784 | 5.826 ± 0.442 |
1.203 ± 0.173 | 3.171 ± 0.442 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
