
Humibacillus xanthopallidus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Intrasporangiaceae; Humibacillus
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
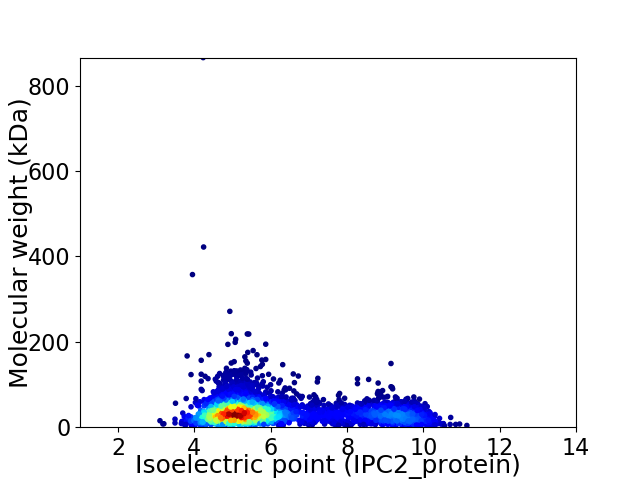
Virtual 2D-PAGE plot for 4527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A543H9Y6|A0A543H9Y6_9MICO Short subunit dehydrogenase OS=Humibacillus xanthopallidus OX=412689 GN=FBY41_4484 PE=4 SV=1
MM1 pKa = 7.19SRR3 pKa = 11.84VSALLSRR10 pKa = 11.84IAAACAVLLVAALLAPAGSAAAAPVAGEE38 pKa = 3.91MSYY41 pKa = 11.6ACALKK46 pKa = 10.98SNGVLRR52 pKa = 11.84AVSSLSEE59 pKa = 4.18CKK61 pKa = 10.7GNEE64 pKa = 4.01TKK66 pKa = 9.48VTLKK70 pKa = 10.21PGPAIFCVQPSGSTRR85 pKa = 11.84LATKK89 pKa = 10.3PKK91 pKa = 9.68DD92 pKa = 3.89CKK94 pKa = 11.07SPATALTLPPTSGTVYY110 pKa = 10.12FCAALPSGTLRR121 pKa = 11.84YY122 pKa = 8.61VTGPGQCLTGEE133 pKa = 4.44VQVQVTPNDD142 pKa = 3.58AAPSVTSTSPADD154 pKa = 3.74GATHH158 pKa = 6.54VATDD162 pKa = 3.35VSPTVTFSEE171 pKa = 4.74PVTATVGSFAFACDD185 pKa = 3.32ATPIPFAVSGSGGSTLTLDD204 pKa = 3.9PTGSLPQGASCTVTVYY220 pKa = 10.92AAGVTDD226 pKa = 4.75VDD228 pKa = 5.25SLDD231 pKa = 4.2PPDD234 pKa = 4.56NMVANRR240 pKa = 11.84TFSFEE245 pKa = 3.58TDD247 pKa = 3.16AAPSLVSSTPVDD259 pKa = 3.6GAVGVATSTNIVLTFSEE276 pKa = 4.71PVDD279 pKa = 3.69VANGAFSLACGGPALPYY296 pKa = 9.58TVTGSGTSTVTVNPDD311 pKa = 2.69ADD313 pKa = 3.91LAQTATCTLTAPAADD328 pKa = 3.68ITDD331 pKa = 3.59VDD333 pKa = 5.1AGDD336 pKa = 4.22PPNALTAAVDD346 pKa = 3.43ISFQTVDD353 pKa = 3.29AAPTLVSISPADD365 pKa = 3.71GATHH369 pKa = 6.63VATDD373 pKa = 3.82ANIVMTFSEE382 pKa = 4.81PVSVPNAGTFSLEE395 pKa = 4.22CPAGFPQGYY404 pKa = 6.95TVLGLSGTTLTIDD417 pKa = 3.53PAGPLPEE424 pKa = 6.12GEE426 pKa = 4.39TCRR429 pKa = 11.84FTGDD433 pKa = 3.17ASTITDD439 pKa = 3.95NDD441 pKa = 3.77AVDD444 pKa = 4.23PPDD447 pKa = 4.18NLTANIDD454 pKa = 3.22ASFTVDD460 pKa = 3.24SPPAVVSTDD469 pKa = 3.43PADD472 pKa = 3.57NATNVPADD480 pKa = 3.56QEE482 pKa = 4.39LTVTFSEE489 pKa = 4.8PVTVSQSSFTLTCDD503 pKa = 3.35SNPVAFTIGGTDD515 pKa = 3.32TAVTLTPTSALPGTAACLLTVVAAQVSDD543 pKa = 4.14ADD545 pKa = 4.46AGDD548 pKa = 4.47PPDD551 pKa = 3.74TMTANVTVSFTTVDD565 pKa = 3.38TAPTVVSTSPTNGATDD581 pKa = 3.6VASGATITVTFSEE594 pKa = 5.18PVTADD599 pKa = 2.84AGAFSLEE606 pKa = 4.41CPVATTTAFALSGSGTDD623 pKa = 3.16WTLTPDD629 pKa = 3.74AKK631 pKa = 10.91LPAAVLCTLTIDD643 pKa = 3.58GSKK646 pKa = 9.61IHH648 pKa = 7.02DD649 pKa = 4.35TDD651 pKa = 3.79TVDD654 pKa = 4.72PPDD657 pKa = 3.78EE658 pKa = 4.19MVADD662 pKa = 3.74VTVEE666 pKa = 3.74FTVAANSAPTDD677 pKa = 3.81LALSAATVAEE687 pKa = 4.15NQPSGTAVGTFSSTDD702 pKa = 3.68PDD704 pKa = 4.14PGDD707 pKa = 3.47TFTYY711 pKa = 10.18TLDD714 pKa = 4.05AGPGDD719 pKa = 3.88TDD721 pKa = 3.3NGSFTIVGDD730 pKa = 3.91KK731 pKa = 10.8LRR733 pKa = 11.84TAASFDD739 pKa = 3.94FEE741 pKa = 5.14AKK743 pKa = 10.02ASYY746 pKa = 9.91SINVRR751 pKa = 11.84TTDD754 pKa = 2.86SAGNVFEE761 pKa = 4.43KK762 pKa = 10.95SFVITVTDD770 pKa = 3.66VNEE773 pKa = 4.19APTDD777 pKa = 3.47ISLSNATVAEE787 pKa = 4.26NEE789 pKa = 3.95PAATVVGSLTATDD802 pKa = 3.87PDD804 pKa = 3.77AGQSHH809 pKa = 6.85TFTVVTTGCGGTYY822 pKa = 10.61ADD824 pKa = 4.2GSSFTVTGGSLVTSAPLDD842 pKa = 3.79YY843 pKa = 9.98EE844 pKa = 4.49AKK846 pKa = 10.19SSYY849 pKa = 7.43TVCVRR854 pKa = 11.84VTDD857 pKa = 3.48SGTPGLTFDD866 pKa = 4.16EE867 pKa = 5.37LFTITVTNVNEE878 pKa = 4.04APVVGGDD885 pKa = 3.83SYY887 pKa = 11.99SGAIGNTLALRR898 pKa = 11.84GVTATGPSTTLTGALPLANDD918 pKa = 4.09TDD920 pKa = 4.38PEE922 pKa = 5.78GDD924 pKa = 3.87TISVVAGTLATTGGGSVTINANGTFSYY951 pKa = 10.68LPGVGDD957 pKa = 3.66VGQTDD962 pKa = 3.91TVTYY966 pKa = 9.47TVTDD970 pKa = 3.84GLLTSTGTMSIAIGNDD986 pKa = 3.02RR987 pKa = 11.84VWWVNAAAPAGGDD1000 pKa = 3.58GRR1002 pKa = 11.84STAPLTTLTTLNGAGGSGDD1021 pKa = 3.16VDD1023 pKa = 3.71GPGDD1027 pKa = 3.72YY1028 pKa = 11.08LFVYY1032 pKa = 10.01AAAGSYY1038 pKa = 10.58AGGLALEE1045 pKa = 4.68ANQRR1049 pKa = 11.84LHH1051 pKa = 5.52GQRR1054 pKa = 11.84HH1055 pKa = 5.6GLTVGGTVLVPAGATAPTITNAAGNGVTLANGVDD1089 pKa = 3.83VQGVTVSGASADD1101 pKa = 4.41GIHH1104 pKa = 6.93GDD1106 pKa = 3.59NVTTATIGSAVTVSGSGADD1125 pKa = 4.38GIDD1128 pKa = 3.33LTGAATGSIGVAATVTGSTARR1149 pKa = 11.84SVNVSNRR1156 pKa = 11.84SGGTTSFTGPISGTGINLASNTGATVTFSAALTISSGGTSAFSATGGGTVTASDD1210 pKa = 3.71TTSTLTSTTGSTLVVEE1226 pKa = 4.43NTTIGGAGLKK1236 pKa = 9.88FRR1238 pKa = 11.84SISANGAANGIRR1250 pKa = 11.84LNTTGSSGGLSVLGGGNTSLGGNASGGTIQNTTGPGVSLTSTSSVSLNNLTVSNTPGASGIKK1312 pKa = 8.35GTGVAGFSFTYY1323 pKa = 9.05GTVTGSGSAGGGAAGAVASNIAFDD1347 pKa = 3.87GTGDD1351 pKa = 4.21GISNVSGAVTVSNSVLTSGYY1371 pKa = 9.92NHH1373 pKa = 7.48GLTIYY1378 pKa = 10.02NNSGVISTLTVTGNTITSAATAAGSIGSGVLVQEE1412 pKa = 5.04FGSAGTGATITTGSISSNSITGFPNGSGIRR1442 pKa = 11.84LTGGNPTSLAAPVSQIGTSGSHH1464 pKa = 6.45FAITGNLVHH1473 pKa = 6.8GFSTSVLMSTEE1484 pKa = 4.24AIIVLAQGRR1493 pKa = 11.84GTSYY1497 pKa = 11.14IDD1499 pKa = 3.15VTNNGTVANPVGLTQGDD1516 pKa = 4.52GISVNATWQHH1526 pKa = 5.75NLTSAITGNVIAPGTGGVVTNRR1548 pKa = 11.84GITGGAAAGGIPGGTGDD1565 pKa = 3.94TAVLNATISNNTVSQTRR1582 pKa = 11.84GNGIYY1587 pKa = 9.95FVARR1591 pKa = 11.84EE1592 pKa = 4.02SSTLRR1597 pKa = 11.84TTVQNNSVAAPTLIGAGIRR1616 pKa = 11.84FDD1618 pKa = 4.08SGTPAPNSVNATLCALISGNTTAGFSPNPGIALRR1652 pKa = 11.84KK1653 pKa = 9.28EE1654 pKa = 4.25GTVSNVNTFGIFGLSPSPTTTALEE1678 pKa = 4.3TTSYY1682 pKa = 11.37VATQNPASSGGALLVSAQTGFTACTPP1708 pKa = 3.79
MM1 pKa = 7.19SRR3 pKa = 11.84VSALLSRR10 pKa = 11.84IAAACAVLLVAALLAPAGSAAAAPVAGEE38 pKa = 3.91MSYY41 pKa = 11.6ACALKK46 pKa = 10.98SNGVLRR52 pKa = 11.84AVSSLSEE59 pKa = 4.18CKK61 pKa = 10.7GNEE64 pKa = 4.01TKK66 pKa = 9.48VTLKK70 pKa = 10.21PGPAIFCVQPSGSTRR85 pKa = 11.84LATKK89 pKa = 10.3PKK91 pKa = 9.68DD92 pKa = 3.89CKK94 pKa = 11.07SPATALTLPPTSGTVYY110 pKa = 10.12FCAALPSGTLRR121 pKa = 11.84YY122 pKa = 8.61VTGPGQCLTGEE133 pKa = 4.44VQVQVTPNDD142 pKa = 3.58AAPSVTSTSPADD154 pKa = 3.74GATHH158 pKa = 6.54VATDD162 pKa = 3.35VSPTVTFSEE171 pKa = 4.74PVTATVGSFAFACDD185 pKa = 3.32ATPIPFAVSGSGGSTLTLDD204 pKa = 3.9PTGSLPQGASCTVTVYY220 pKa = 10.92AAGVTDD226 pKa = 4.75VDD228 pKa = 5.25SLDD231 pKa = 4.2PPDD234 pKa = 4.56NMVANRR240 pKa = 11.84TFSFEE245 pKa = 3.58TDD247 pKa = 3.16AAPSLVSSTPVDD259 pKa = 3.6GAVGVATSTNIVLTFSEE276 pKa = 4.71PVDD279 pKa = 3.69VANGAFSLACGGPALPYY296 pKa = 9.58TVTGSGTSTVTVNPDD311 pKa = 2.69ADD313 pKa = 3.91LAQTATCTLTAPAADD328 pKa = 3.68ITDD331 pKa = 3.59VDD333 pKa = 5.1AGDD336 pKa = 4.22PPNALTAAVDD346 pKa = 3.43ISFQTVDD353 pKa = 3.29AAPTLVSISPADD365 pKa = 3.71GATHH369 pKa = 6.63VATDD373 pKa = 3.82ANIVMTFSEE382 pKa = 4.81PVSVPNAGTFSLEE395 pKa = 4.22CPAGFPQGYY404 pKa = 6.95TVLGLSGTTLTIDD417 pKa = 3.53PAGPLPEE424 pKa = 6.12GEE426 pKa = 4.39TCRR429 pKa = 11.84FTGDD433 pKa = 3.17ASTITDD439 pKa = 3.95NDD441 pKa = 3.77AVDD444 pKa = 4.23PPDD447 pKa = 4.18NLTANIDD454 pKa = 3.22ASFTVDD460 pKa = 3.24SPPAVVSTDD469 pKa = 3.43PADD472 pKa = 3.57NATNVPADD480 pKa = 3.56QEE482 pKa = 4.39LTVTFSEE489 pKa = 4.8PVTVSQSSFTLTCDD503 pKa = 3.35SNPVAFTIGGTDD515 pKa = 3.32TAVTLTPTSALPGTAACLLTVVAAQVSDD543 pKa = 4.14ADD545 pKa = 4.46AGDD548 pKa = 4.47PPDD551 pKa = 3.74TMTANVTVSFTTVDD565 pKa = 3.38TAPTVVSTSPTNGATDD581 pKa = 3.6VASGATITVTFSEE594 pKa = 5.18PVTADD599 pKa = 2.84AGAFSLEE606 pKa = 4.41CPVATTTAFALSGSGTDD623 pKa = 3.16WTLTPDD629 pKa = 3.74AKK631 pKa = 10.91LPAAVLCTLTIDD643 pKa = 3.58GSKK646 pKa = 9.61IHH648 pKa = 7.02DD649 pKa = 4.35TDD651 pKa = 3.79TVDD654 pKa = 4.72PPDD657 pKa = 3.78EE658 pKa = 4.19MVADD662 pKa = 3.74VTVEE666 pKa = 3.74FTVAANSAPTDD677 pKa = 3.81LALSAATVAEE687 pKa = 4.15NQPSGTAVGTFSSTDD702 pKa = 3.68PDD704 pKa = 4.14PGDD707 pKa = 3.47TFTYY711 pKa = 10.18TLDD714 pKa = 4.05AGPGDD719 pKa = 3.88TDD721 pKa = 3.3NGSFTIVGDD730 pKa = 3.91KK731 pKa = 10.8LRR733 pKa = 11.84TAASFDD739 pKa = 3.94FEE741 pKa = 5.14AKK743 pKa = 10.02ASYY746 pKa = 9.91SINVRR751 pKa = 11.84TTDD754 pKa = 2.86SAGNVFEE761 pKa = 4.43KK762 pKa = 10.95SFVITVTDD770 pKa = 3.66VNEE773 pKa = 4.19APTDD777 pKa = 3.47ISLSNATVAEE787 pKa = 4.26NEE789 pKa = 3.95PAATVVGSLTATDD802 pKa = 3.87PDD804 pKa = 3.77AGQSHH809 pKa = 6.85TFTVVTTGCGGTYY822 pKa = 10.61ADD824 pKa = 4.2GSSFTVTGGSLVTSAPLDD842 pKa = 3.79YY843 pKa = 9.98EE844 pKa = 4.49AKK846 pKa = 10.19SSYY849 pKa = 7.43TVCVRR854 pKa = 11.84VTDD857 pKa = 3.48SGTPGLTFDD866 pKa = 4.16EE867 pKa = 5.37LFTITVTNVNEE878 pKa = 4.04APVVGGDD885 pKa = 3.83SYY887 pKa = 11.99SGAIGNTLALRR898 pKa = 11.84GVTATGPSTTLTGALPLANDD918 pKa = 4.09TDD920 pKa = 4.38PEE922 pKa = 5.78GDD924 pKa = 3.87TISVVAGTLATTGGGSVTINANGTFSYY951 pKa = 10.68LPGVGDD957 pKa = 3.66VGQTDD962 pKa = 3.91TVTYY966 pKa = 9.47TVTDD970 pKa = 3.84GLLTSTGTMSIAIGNDD986 pKa = 3.02RR987 pKa = 11.84VWWVNAAAPAGGDD1000 pKa = 3.58GRR1002 pKa = 11.84STAPLTTLTTLNGAGGSGDD1021 pKa = 3.16VDD1023 pKa = 3.71GPGDD1027 pKa = 3.72YY1028 pKa = 11.08LFVYY1032 pKa = 10.01AAAGSYY1038 pKa = 10.58AGGLALEE1045 pKa = 4.68ANQRR1049 pKa = 11.84LHH1051 pKa = 5.52GQRR1054 pKa = 11.84HH1055 pKa = 5.6GLTVGGTVLVPAGATAPTITNAAGNGVTLANGVDD1089 pKa = 3.83VQGVTVSGASADD1101 pKa = 4.41GIHH1104 pKa = 6.93GDD1106 pKa = 3.59NVTTATIGSAVTVSGSGADD1125 pKa = 4.38GIDD1128 pKa = 3.33LTGAATGSIGVAATVTGSTARR1149 pKa = 11.84SVNVSNRR1156 pKa = 11.84SGGTTSFTGPISGTGINLASNTGATVTFSAALTISSGGTSAFSATGGGTVTASDD1210 pKa = 3.71TTSTLTSTTGSTLVVEE1226 pKa = 4.43NTTIGGAGLKK1236 pKa = 9.88FRR1238 pKa = 11.84SISANGAANGIRR1250 pKa = 11.84LNTTGSSGGLSVLGGGNTSLGGNASGGTIQNTTGPGVSLTSTSSVSLNNLTVSNTPGASGIKK1312 pKa = 8.35GTGVAGFSFTYY1323 pKa = 9.05GTVTGSGSAGGGAAGAVASNIAFDD1347 pKa = 3.87GTGDD1351 pKa = 4.21GISNVSGAVTVSNSVLTSGYY1371 pKa = 9.92NHH1373 pKa = 7.48GLTIYY1378 pKa = 10.02NNSGVISTLTVTGNTITSAATAAGSIGSGVLVQEE1412 pKa = 5.04FGSAGTGATITTGSISSNSITGFPNGSGIRR1442 pKa = 11.84LTGGNPTSLAAPVSQIGTSGSHH1464 pKa = 6.45FAITGNLVHH1473 pKa = 6.8GFSTSVLMSTEE1484 pKa = 4.24AIIVLAQGRR1493 pKa = 11.84GTSYY1497 pKa = 11.14IDD1499 pKa = 3.15VTNNGTVANPVGLTQGDD1516 pKa = 4.52GISVNATWQHH1526 pKa = 5.75NLTSAITGNVIAPGTGGVVTNRR1548 pKa = 11.84GITGGAAAGGIPGGTGDD1565 pKa = 3.94TAVLNATISNNTVSQTRR1582 pKa = 11.84GNGIYY1587 pKa = 9.95FVARR1591 pKa = 11.84EE1592 pKa = 4.02SSTLRR1597 pKa = 11.84TTVQNNSVAAPTLIGAGIRR1616 pKa = 11.84FDD1618 pKa = 4.08SGTPAPNSVNATLCALISGNTTAGFSPNPGIALRR1652 pKa = 11.84KK1653 pKa = 9.28EE1654 pKa = 4.25GTVSNVNTFGIFGLSPSPTTTALEE1678 pKa = 4.3TTSYY1682 pKa = 11.37VATQNPASSGGALLVSAQTGFTACTPP1708 pKa = 3.79
Molecular weight: 166.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A543H8F6|A0A543H8F6_9MICO Polyketide cyclase/dehydrase/lipid transport protein OS=Humibacillus xanthopallidus OX=412689 GN=FBY41_4617 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1567071 |
29 |
8685 |
346.2 |
36.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.34 ± 0.049 | 0.675 ± 0.009 |
6.316 ± 0.032 | 5.309 ± 0.043 |
2.733 ± 0.025 | 9.297 ± 0.04 |
2.252 ± 0.019 | 3.552 ± 0.029 |
1.852 ± 0.027 | 10.027 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.759 ± 0.017 | 1.748 ± 0.027 |
5.82 ± 0.029 | 2.717 ± 0.019 |
7.552 ± 0.054 | 5.696 ± 0.033 |
6.505 ± 0.057 | 9.364 ± 0.036 |
1.606 ± 0.016 | 1.878 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
