
Thrips-associated genomovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus echiam1
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
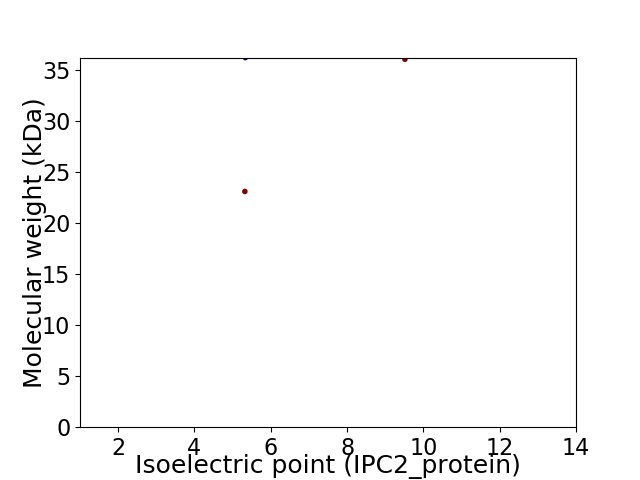
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8YT70|A0A1P8YT70_9VIRU RepA OS=Thrips-associated genomovirus 1 OX=1941233 PE=3 SV=1
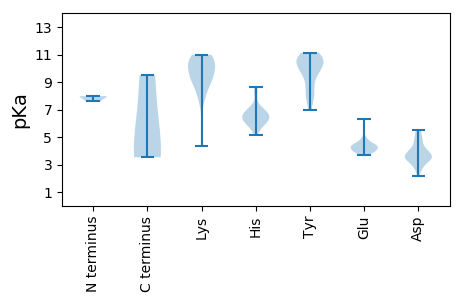
MM1 pKa = 7.97PPFTYY6 pKa = 9.91NARR9 pKa = 11.84YY10 pKa = 9.11FLVTYY15 pKa = 7.74AQCGDD20 pKa = 3.98LDD22 pKa = 3.6PWAVNDD28 pKa = 4.06HH29 pKa = 6.05FASLGAEE36 pKa = 3.95CIVARR41 pKa = 11.84EE42 pKa = 4.04SHH44 pKa = 6.5ADD46 pKa = 3.41EE47 pKa = 4.79GTHH50 pKa = 6.18LHH52 pKa = 6.43AFVDD56 pKa = 4.82FGRR59 pKa = 11.84KK60 pKa = 8.17FRR62 pKa = 11.84SRR64 pKa = 11.84NARR67 pKa = 11.84KK68 pKa = 9.06FDD70 pKa = 3.3VDD72 pKa = 3.22GRR74 pKa = 11.84HH75 pKa = 6.35ANIAPSKK82 pKa = 9.24GKK84 pKa = 9.72PEE86 pKa = 3.91GGYY89 pKa = 10.69DD90 pKa = 3.55YY91 pKa = 10.74AIKK94 pKa = 10.94DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PSVNGAVSNGDD116 pKa = 2.92KK117 pKa = 10.14WSEE120 pKa = 3.78IVGAEE125 pKa = 3.99SEE127 pKa = 4.41QLFWEE132 pKa = 4.43AVEE135 pKa = 4.52RR136 pKa = 11.84LDD138 pKa = 5.48PKK140 pKa = 10.91ALCTNYY146 pKa = 10.69GNLRR150 pKa = 11.84KK151 pKa = 9.28FADD154 pKa = 2.71WRR156 pKa = 11.84YY157 pKa = 8.5RR158 pKa = 11.84VEE160 pKa = 4.02PEE162 pKa = 4.24RR163 pKa = 11.84YY164 pKa = 8.04EE165 pKa = 4.33HH166 pKa = 6.82PAGLNFEE173 pKa = 4.91LGMVPDD179 pKa = 4.23LAAWGGFGSGDD190 pKa = 3.45DD191 pKa = 3.82TVGGRR196 pKa = 11.84VRR198 pKa = 11.84SLCLYY203 pKa = 10.7GPTKK207 pKa = 10.49VGKK210 pKa = 5.85TTWARR215 pKa = 11.84SLGNHH220 pKa = 5.79VYY222 pKa = 10.79FMGLMSGEE230 pKa = 4.06VALRR234 pKa = 11.84DD235 pKa = 3.82MPDD238 pKa = 2.72AEE240 pKa = 4.32YY241 pKa = 11.01AVFDD245 pKa = 4.67DD246 pKa = 3.76MRR248 pKa = 11.84GGIKK252 pKa = 10.28FFQAWKK258 pKa = 9.36EE259 pKa = 3.87WFGAQLVVTVKK270 pKa = 10.52KK271 pKa = 10.68LYY273 pKa = 10.05RR274 pKa = 11.84DD275 pKa = 3.54PVQVKK280 pKa = 8.69WGKK283 pKa = 9.01PCIWLCNEE291 pKa = 4.51DD292 pKa = 3.92PRR294 pKa = 11.84TSLEE298 pKa = 3.72PAEE301 pKa = 4.57IDD303 pKa = 3.53WIEE306 pKa = 4.14GNCDD310 pKa = 3.96FIDD313 pKa = 3.57VHH315 pKa = 8.16SSIISHH321 pKa = 6.95ANTT324 pKa = 3.53
MM1 pKa = 7.97PPFTYY6 pKa = 9.91NARR9 pKa = 11.84YY10 pKa = 9.11FLVTYY15 pKa = 7.74AQCGDD20 pKa = 3.98LDD22 pKa = 3.6PWAVNDD28 pKa = 4.06HH29 pKa = 6.05FASLGAEE36 pKa = 3.95CIVARR41 pKa = 11.84EE42 pKa = 4.04SHH44 pKa = 6.5ADD46 pKa = 3.41EE47 pKa = 4.79GTHH50 pKa = 6.18LHH52 pKa = 6.43AFVDD56 pKa = 4.82FGRR59 pKa = 11.84KK60 pKa = 8.17FRR62 pKa = 11.84SRR64 pKa = 11.84NARR67 pKa = 11.84KK68 pKa = 9.06FDD70 pKa = 3.3VDD72 pKa = 3.22GRR74 pKa = 11.84HH75 pKa = 6.35ANIAPSKK82 pKa = 9.24GKK84 pKa = 9.72PEE86 pKa = 3.91GGYY89 pKa = 10.69DD90 pKa = 3.55YY91 pKa = 10.74AIKK94 pKa = 10.94DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PSVNGAVSNGDD116 pKa = 2.92KK117 pKa = 10.14WSEE120 pKa = 3.78IVGAEE125 pKa = 3.99SEE127 pKa = 4.41QLFWEE132 pKa = 4.43AVEE135 pKa = 4.52RR136 pKa = 11.84LDD138 pKa = 5.48PKK140 pKa = 10.91ALCTNYY146 pKa = 10.69GNLRR150 pKa = 11.84KK151 pKa = 9.28FADD154 pKa = 2.71WRR156 pKa = 11.84YY157 pKa = 8.5RR158 pKa = 11.84VEE160 pKa = 4.02PEE162 pKa = 4.24RR163 pKa = 11.84YY164 pKa = 8.04EE165 pKa = 4.33HH166 pKa = 6.82PAGLNFEE173 pKa = 4.91LGMVPDD179 pKa = 4.23LAAWGGFGSGDD190 pKa = 3.45DD191 pKa = 3.82TVGGRR196 pKa = 11.84VRR198 pKa = 11.84SLCLYY203 pKa = 10.7GPTKK207 pKa = 10.49VGKK210 pKa = 5.85TTWARR215 pKa = 11.84SLGNHH220 pKa = 5.79VYY222 pKa = 10.79FMGLMSGEE230 pKa = 4.06VALRR234 pKa = 11.84DD235 pKa = 3.82MPDD238 pKa = 2.72AEE240 pKa = 4.32YY241 pKa = 11.01AVFDD245 pKa = 4.67DD246 pKa = 3.76MRR248 pKa = 11.84GGIKK252 pKa = 10.28FFQAWKK258 pKa = 9.36EE259 pKa = 3.87WFGAQLVVTVKK270 pKa = 10.52KK271 pKa = 10.68LYY273 pKa = 10.05RR274 pKa = 11.84DD275 pKa = 3.54PVQVKK280 pKa = 8.69WGKK283 pKa = 9.01PCIWLCNEE291 pKa = 4.51DD292 pKa = 3.92PRR294 pKa = 11.84TSLEE298 pKa = 3.72PAEE301 pKa = 4.57IDD303 pKa = 3.53WIEE306 pKa = 4.14GNCDD310 pKa = 3.96FIDD313 pKa = 3.57VHH315 pKa = 8.16SSIISHH321 pKa = 6.95ANTT324 pKa = 3.53
Molecular weight: 36.2 kDa
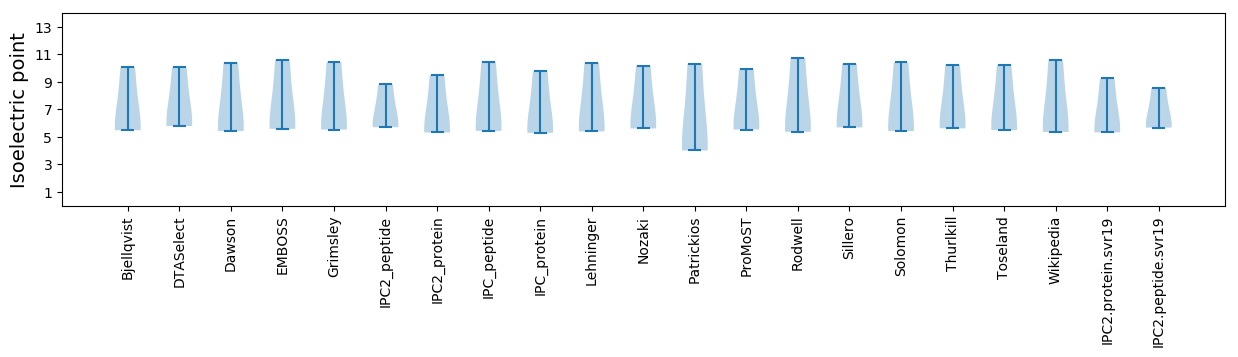
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8YT68|A0A1P8YT68_9VIRU Replication-associated protein OS=Thrips-associated genomovirus 1 OX=1941233 PE=3 SV=1
MM1 pKa = 7.6AYY3 pKa = 10.17ARR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 9.77GSYY11 pKa = 9.84RR12 pKa = 11.84AKK14 pKa = 9.83KK15 pKa = 4.35TTKK18 pKa = 9.67RR19 pKa = 11.84SGRR22 pKa = 11.84RR23 pKa = 11.84SPYY26 pKa = 6.97TAKK29 pKa = 9.5TRR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.75KK34 pKa = 10.55KK35 pKa = 8.02RR36 pKa = 11.84TYY38 pKa = 10.45RR39 pKa = 11.84KK40 pKa = 7.96RR41 pKa = 11.84TMSTKK46 pKa = 10.53SILNKK51 pKa = 10.02SSRR54 pKa = 11.84KK55 pKa = 9.13KK56 pKa = 10.28RR57 pKa = 11.84NGMLTFSNSTSSGALATVAQNPLTIAGSPSGDD89 pKa = 3.5TIGLVHH95 pKa = 6.83FRR97 pKa = 11.84PTAMDD102 pKa = 4.17LDD104 pKa = 4.69DD105 pKa = 5.5NVTNPNSIVATPSRR119 pKa = 11.84TATVCFMRR127 pKa = 11.84GFLEE131 pKa = 4.38NIRR134 pKa = 11.84VEE136 pKa = 4.41TSSGNPWFHH145 pKa = 6.83RR146 pKa = 11.84RR147 pKa = 11.84VCICSRR153 pKa = 11.84DD154 pKa = 3.64PRR156 pKa = 11.84LLLLSSSDD164 pKa = 3.25TTGVDD169 pKa = 2.2RR170 pKa = 11.84GYY172 pKa = 10.78AASGSIEE179 pKa = 4.24TSNGWQRR186 pKa = 11.84LASNVLTDD194 pKa = 3.41TMTNTTTNWLGLLFKK209 pKa = 10.55GAQGVDD215 pKa = 2.56WDD217 pKa = 4.99DD218 pKa = 4.9YY219 pKa = 10.03ITAPVDD225 pKa = 3.35TTRR228 pKa = 11.84VDD230 pKa = 3.78LKK232 pKa = 10.99FDD234 pKa = 3.36KK235 pKa = 10.49TWIYY239 pKa = 11.12KK240 pKa = 10.19SGNQQGILKK249 pKa = 7.55EE250 pKa = 4.35TKK252 pKa = 9.08LWHH255 pKa = 5.79PMNRR259 pKa = 11.84NLYY262 pKa = 10.35YY263 pKa = 10.76DD264 pKa = 4.47DD265 pKa = 5.48DD266 pKa = 4.72EE267 pKa = 6.32NGAGEE272 pKa = 4.23TTRR275 pKa = 11.84NFSVQDD281 pKa = 3.29KK282 pKa = 10.75RR283 pKa = 11.84GMGDD287 pKa = 2.91YY288 pKa = 10.63HH289 pKa = 8.66IFDD292 pKa = 5.17LFSQGSSGSTADD304 pKa = 3.48RR305 pKa = 11.84LKK307 pKa = 10.67IRR309 pKa = 11.84FTSTMYY315 pKa = 9.45WHH317 pKa = 7.08EE318 pKa = 4.17KK319 pKa = 9.5
MM1 pKa = 7.6AYY3 pKa = 10.17ARR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 9.77GSYY11 pKa = 9.84RR12 pKa = 11.84AKK14 pKa = 9.83KK15 pKa = 4.35TTKK18 pKa = 9.67RR19 pKa = 11.84SGRR22 pKa = 11.84RR23 pKa = 11.84SPYY26 pKa = 6.97TAKK29 pKa = 9.5TRR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.75KK34 pKa = 10.55KK35 pKa = 8.02RR36 pKa = 11.84TYY38 pKa = 10.45RR39 pKa = 11.84KK40 pKa = 7.96RR41 pKa = 11.84TMSTKK46 pKa = 10.53SILNKK51 pKa = 10.02SSRR54 pKa = 11.84KK55 pKa = 9.13KK56 pKa = 10.28RR57 pKa = 11.84NGMLTFSNSTSSGALATVAQNPLTIAGSPSGDD89 pKa = 3.5TIGLVHH95 pKa = 6.83FRR97 pKa = 11.84PTAMDD102 pKa = 4.17LDD104 pKa = 4.69DD105 pKa = 5.5NVTNPNSIVATPSRR119 pKa = 11.84TATVCFMRR127 pKa = 11.84GFLEE131 pKa = 4.38NIRR134 pKa = 11.84VEE136 pKa = 4.41TSSGNPWFHH145 pKa = 6.83RR146 pKa = 11.84RR147 pKa = 11.84VCICSRR153 pKa = 11.84DD154 pKa = 3.64PRR156 pKa = 11.84LLLLSSSDD164 pKa = 3.25TTGVDD169 pKa = 2.2RR170 pKa = 11.84GYY172 pKa = 10.78AASGSIEE179 pKa = 4.24TSNGWQRR186 pKa = 11.84LASNVLTDD194 pKa = 3.41TMTNTTTNWLGLLFKK209 pKa = 10.55GAQGVDD215 pKa = 2.56WDD217 pKa = 4.99DD218 pKa = 4.9YY219 pKa = 10.03ITAPVDD225 pKa = 3.35TTRR228 pKa = 11.84VDD230 pKa = 3.78LKK232 pKa = 10.99FDD234 pKa = 3.36KK235 pKa = 10.49TWIYY239 pKa = 11.12KK240 pKa = 10.19SGNQQGILKK249 pKa = 7.55EE250 pKa = 4.35TKK252 pKa = 9.08LWHH255 pKa = 5.79PMNRR259 pKa = 11.84NLYY262 pKa = 10.35YY263 pKa = 10.76DD264 pKa = 4.47DD265 pKa = 5.48DD266 pKa = 4.72EE267 pKa = 6.32NGAGEE272 pKa = 4.23TTRR275 pKa = 11.84NFSVQDD281 pKa = 3.29KK282 pKa = 10.75RR283 pKa = 11.84GMGDD287 pKa = 2.91YY288 pKa = 10.63HH289 pKa = 8.66IFDD292 pKa = 5.17LFSQGSSGSTADD304 pKa = 3.48RR305 pKa = 11.84LKK307 pKa = 10.67IRR309 pKa = 11.84FTSTMYY315 pKa = 9.45WHH317 pKa = 7.08EE318 pKa = 4.17KK319 pKa = 9.5
Molecular weight: 36.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
853 |
210 |
324 |
284.3 |
31.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.441 ± 1.237 | 1.524 ± 0.326 |
7.268 ± 0.307 | 4.807 ± 1.137 |
4.572 ± 0.496 | 9.613 ± 0.928 |
2.462 ± 0.391 | 3.165 ± 0.377 |
5.158 ± 0.506 | 6.682 ± 0.109 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.993 ± 0.381 | 4.689 ± 0.435 |
4.455 ± 0.575 | 1.641 ± 0.254 |
7.62 ± 0.85 | 6.8 ± 1.418 |
6.213 ± 2.344 | 6.213 ± 0.973 |
2.814 ± 0.316 | 3.869 ± 0.098 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
