
Ophiocordyceps camponoti-leonardi (nom. inval.)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Ophiocordycipitaceae; Ophiocordyceps; unclassified Ophiocordyceps
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
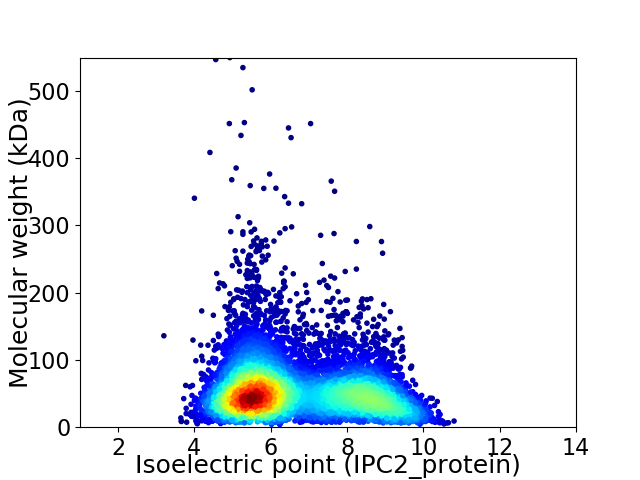
Virtual 2D-PAGE plot for 7043 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A369GQ40|A0A369GQ40_9HYPO Uncharacterized protein OS=Ophiocordyceps camponoti-leonardi (nom. inval.) OX=2039875 GN=CP532_1056 PE=4 SV=1
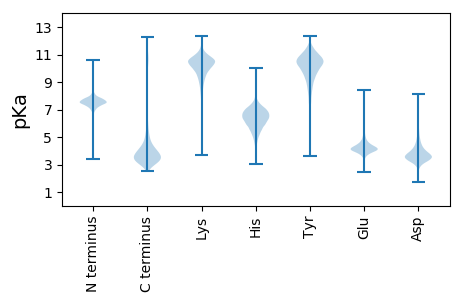
MM1 pKa = 7.16VSWEE5 pKa = 4.29RR6 pKa = 11.84DD7 pKa = 3.35FKK9 pKa = 11.0MEE11 pKa = 4.16EE12 pKa = 3.35VWYY15 pKa = 9.98VCGVCGLEE23 pKa = 4.99HH24 pKa = 7.96DD25 pKa = 4.68DD26 pKa = 3.67AQHH29 pKa = 6.67RR30 pKa = 11.84STYY33 pKa = 9.78RR34 pKa = 11.84CYY36 pKa = 10.9VDD38 pKa = 5.33HH39 pKa = 7.37FNDD42 pKa = 5.77DD43 pKa = 4.99DD44 pKa = 7.09DD45 pKa = 7.56DD46 pKa = 7.2DD47 pKa = 7.56DD48 pKa = 7.55DD49 pKa = 7.23DD50 pKa = 5.91DD51 pKa = 4.25VHH53 pKa = 7.18NNHH56 pKa = 6.46NDD58 pKa = 3.55DD59 pKa = 5.3GDD61 pKa = 5.17DD62 pKa = 4.77DD63 pKa = 5.73DD64 pKa = 5.83EE65 pKa = 5.33NSNNDD70 pKa = 3.12EE71 pKa = 4.08
MM1 pKa = 7.16VSWEE5 pKa = 4.29RR6 pKa = 11.84DD7 pKa = 3.35FKK9 pKa = 11.0MEE11 pKa = 4.16EE12 pKa = 3.35VWYY15 pKa = 9.98VCGVCGLEE23 pKa = 4.99HH24 pKa = 7.96DD25 pKa = 4.68DD26 pKa = 3.67AQHH29 pKa = 6.67RR30 pKa = 11.84STYY33 pKa = 9.78RR34 pKa = 11.84CYY36 pKa = 10.9VDD38 pKa = 5.33HH39 pKa = 7.37FNDD42 pKa = 5.77DD43 pKa = 4.99DD44 pKa = 7.09DD45 pKa = 7.56DD46 pKa = 7.2DD47 pKa = 7.56DD48 pKa = 7.55DD49 pKa = 7.23DD50 pKa = 5.91DD51 pKa = 4.25VHH53 pKa = 7.18NNHH56 pKa = 6.46NDD58 pKa = 3.55DD59 pKa = 5.3GDD61 pKa = 5.17DD62 pKa = 4.77DD63 pKa = 5.73DD64 pKa = 5.83EE65 pKa = 5.33NSNNDD70 pKa = 3.12EE71 pKa = 4.08
Molecular weight: 8.46 kDa
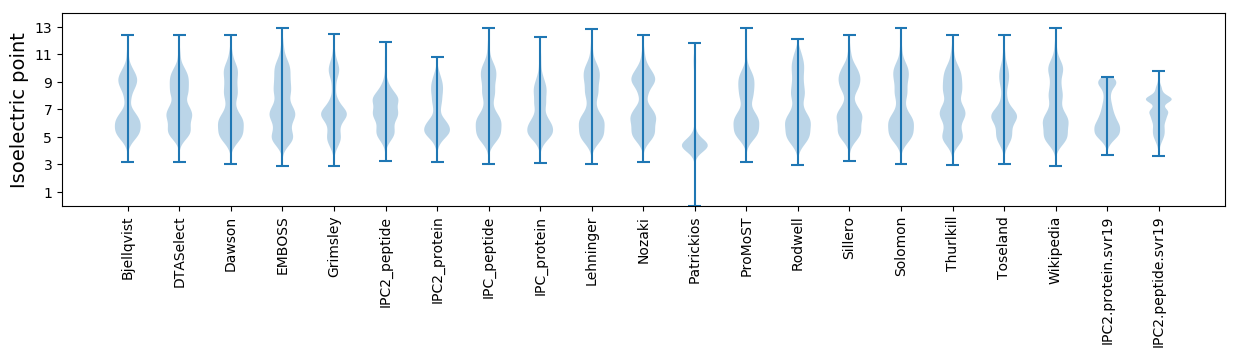
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A369GHF9|A0A369GHF9_9HYPO ATP-dependent (S)-NAD(P)H-hydrate dehydratase OS=Ophiocordyceps camponoti-leonardi (nom. inval.) OX=2039875 GN=CP532_5202 PE=3 SV=1
DDD2 pKa = 3.39KK3 pKa = 10.82KK4 pKa = 10.54KK5 pKa = 10.54KKK7 pKa = 9.16KKK9 pKa = 9.83KK10 pKa = 9.21SLLASILIILRR21 pKa = 11.84TSLEEE26 pKa = 3.95GRR28 pKa = 11.84FPAFCALLVGGSSLLHHH45 pKa = 6.45KK46 pKa = 10.21LTTIVTRR53 pKa = 11.84VARR56 pKa = 11.84PRR58 pKa = 11.84LSNAACLSLARR69 pKa = 11.84WLASFVASWLSLGLLQKKK87 pKa = 10.35RR88 pKa = 11.84PSPLRR93 pKa = 11.84RR94 pKa = 11.84EEE96 pKa = 4.23EE97 pKa = 4.33PPASSGLVPNKKK109 pKa = 10.16DD110 pKa = 3.79DD111 pKa = 3.27EE112 pKa = 4.66MAGKKK117 pKa = 8.14LDDD120 pKa = 3.52TLFAVTQALDDD131 pKa = 3.8IIGEEE136 pKa = 3.97WSRR139 pKa = 11.84RR140 pKa = 11.84KKK142 pKa = 9.81RR143 pKa = 11.84RR144 pKa = 11.84VAARR148 pKa = 11.84KKK150 pKa = 9.03SDDD153 pKa = 3.17DDD155 pKa = 4.51DD156 pKa = 5.26ISRR159 pKa = 11.84MADDD163 pKa = 2.91MSFVASCAFIMWAWFYYY180 pKa = 11.59PDDD183 pKa = 3.94LPRR186 pKa = 11.84SYYY189 pKa = 11.55KKK191 pKa = 9.85ITSAAHHH198 pKa = 5.06DDD200 pKa = 3.65RR201 pKa = 11.84LLEEE205 pKa = 3.91LRR207 pKa = 11.84RR208 pKa = 11.84CRR210 pKa = 11.84TGEEE214 pKa = 3.62RR215 pKa = 11.84YYY217 pKa = 10.22VDDD220 pKa = 2.96GQAPLLGSMCADDD233 pKa = 3.32YY234 pKa = 11.12LPRR237 pKa = 11.84QWGDDD242 pKa = 3.16AVSVPFPCDDD252 pKa = 3.16VHHH255 pKa = 6.82GRR257 pKa = 11.84GPCCEEE263 pKa = 3.6YY264 pKa = 10.47HH265 pKa = 5.13WRR267 pKa = 11.84RR268 pKa = 11.84FSQSWLWSMYYY279 pKa = 10.02YYY281 pKa = 11.12PLAAALQLRR290 pKa = 11.84RR291 pKa = 11.84PTLRR295 pKa = 11.84SPFKKK300 pKa = 10.7VVSAARR306 pKa = 11.84SSAFLATFITLFYYY320 pKa = 11.04YY321 pKa = 8.7VCLGRR326 pKa = 11.84TRR328 pKa = 11.84LGPHHH333 pKa = 6.58LGRR336 pKa = 11.84GPSSCDDD343 pKa = 3.29IDDD346 pKa = 4.68GICVANGCFLCGWSILIEEE365 pKa = 4.17TSRR368 pKa = 11.84RR369 pKa = 11.84KKK371 pKa = 10.68DD372 pKa = 3.26ALFAAPRR379 pKa = 11.84AMATLLPRR387 pKa = 11.84RR388 pKa = 11.84FPTGLQWRR396 pKa = 11.84EEE398 pKa = 3.7LAFAASTAVVFTCVQEEE415 pKa = 3.93PKKK418 pKa = 10.01VRR420 pKa = 11.84GMLGGILSLAMRR432 pKa = 11.84Q
DDD2 pKa = 3.39KK3 pKa = 10.82KK4 pKa = 10.54KK5 pKa = 10.54KKK7 pKa = 9.16KKK9 pKa = 9.83KK10 pKa = 9.21SLLASILIILRR21 pKa = 11.84TSLEEE26 pKa = 3.95GRR28 pKa = 11.84FPAFCALLVGGSSLLHHH45 pKa = 6.45KK46 pKa = 10.21LTTIVTRR53 pKa = 11.84VARR56 pKa = 11.84PRR58 pKa = 11.84LSNAACLSLARR69 pKa = 11.84WLASFVASWLSLGLLQKKK87 pKa = 10.35RR88 pKa = 11.84PSPLRR93 pKa = 11.84RR94 pKa = 11.84EEE96 pKa = 4.23EE97 pKa = 4.33PPASSGLVPNKKK109 pKa = 10.16DD110 pKa = 3.79DD111 pKa = 3.27EE112 pKa = 4.66MAGKKK117 pKa = 8.14LDDD120 pKa = 3.52TLFAVTQALDDD131 pKa = 3.8IIGEEE136 pKa = 3.97WSRR139 pKa = 11.84RR140 pKa = 11.84KKK142 pKa = 9.81RR143 pKa = 11.84RR144 pKa = 11.84VAARR148 pKa = 11.84KKK150 pKa = 9.03SDDD153 pKa = 3.17DDD155 pKa = 4.51DD156 pKa = 5.26ISRR159 pKa = 11.84MADDD163 pKa = 2.91MSFVASCAFIMWAWFYYY180 pKa = 11.59PDDD183 pKa = 3.94LPRR186 pKa = 11.84SYYY189 pKa = 11.55KKK191 pKa = 9.85ITSAAHHH198 pKa = 5.06DDD200 pKa = 3.65RR201 pKa = 11.84LLEEE205 pKa = 3.91LRR207 pKa = 11.84RR208 pKa = 11.84CRR210 pKa = 11.84TGEEE214 pKa = 3.62RR215 pKa = 11.84YYY217 pKa = 10.22VDDD220 pKa = 2.96GQAPLLGSMCADDD233 pKa = 3.32YY234 pKa = 11.12LPRR237 pKa = 11.84QWGDDD242 pKa = 3.16AVSVPFPCDDD252 pKa = 3.16VHHH255 pKa = 6.82GRR257 pKa = 11.84GPCCEEE263 pKa = 3.6YY264 pKa = 10.47HH265 pKa = 5.13WRR267 pKa = 11.84RR268 pKa = 11.84FSQSWLWSMYYY279 pKa = 10.02YYY281 pKa = 11.12PLAAALQLRR290 pKa = 11.84RR291 pKa = 11.84PTLRR295 pKa = 11.84SPFKKK300 pKa = 10.7VVSAARR306 pKa = 11.84SSAFLATFITLFYYY320 pKa = 11.04YY321 pKa = 8.7VCLGRR326 pKa = 11.84TRR328 pKa = 11.84LGPHHH333 pKa = 6.58LGRR336 pKa = 11.84GPSSCDDD343 pKa = 3.29IDDD346 pKa = 4.68GICVANGCFLCGWSILIEEE365 pKa = 4.17TSRR368 pKa = 11.84RR369 pKa = 11.84KKK371 pKa = 10.68DD372 pKa = 3.26ALFAAPRR379 pKa = 11.84AMATLLPRR387 pKa = 11.84RR388 pKa = 11.84FPTGLQWRR396 pKa = 11.84EEE398 pKa = 3.7LAFAASTAVVFTCVQEEE415 pKa = 3.93PKKK418 pKa = 10.01VRR420 pKa = 11.84GMLGGILSLAMRR432 pKa = 11.84Q
Molecular weight: 48.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3884336 |
36 |
5063 |
551.5 |
60.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.399 ± 0.028 | 1.286 ± 0.011 |
6.219 ± 0.021 | 5.99 ± 0.03 |
3.452 ± 0.019 | 7.118 ± 0.038 |
2.4 ± 0.014 | 4.028 ± 0.018 |
4.473 ± 0.024 | 8.977 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.216 ± 0.011 | 3.144 ± 0.016 |
6.196 ± 0.034 | 3.974 ± 0.022 |
7.049 ± 0.025 | 8.345 ± 0.036 |
5.523 ± 0.018 | 6.409 ± 0.02 |
1.373 ± 0.01 | 2.428 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
