
Oleispira antarctica RB-8
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Oleispira; Oleispira antarctica
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
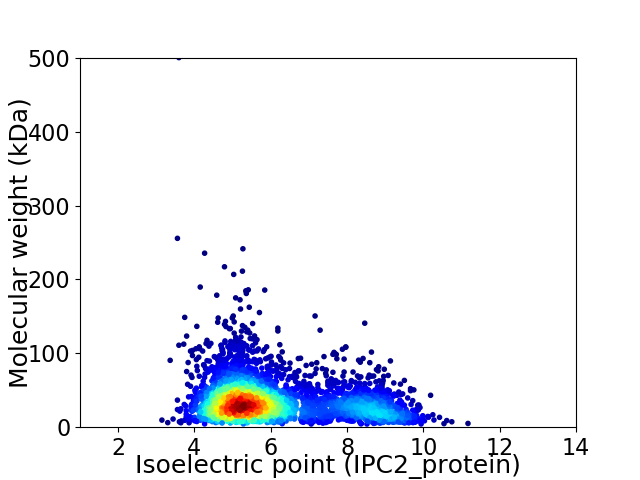
Virtual 2D-PAGE plot for 3895 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R4YPX3|R4YPX3_OLEAN Ethanolamine utilisation protein EutH OS=Oleispira antarctica RB-8 OX=698738 GN=OLEAN_C11370 PE=4 SV=1
MM1 pKa = 7.4KK2 pKa = 10.49SSTPSMKK9 pKa = 10.48LGLASAIAVAALGMTGCGGDD29 pKa = 4.21DD30 pKa = 3.37SSSSSKK36 pKa = 9.55STSNLIDD43 pKa = 3.53PCLNADD49 pKa = 3.78CTNSGFILPSNAIYY63 pKa = 10.78VSADD67 pKa = 3.19AADD70 pKa = 4.07GTDD73 pKa = 4.27LKK75 pKa = 11.15NALITALTDD84 pKa = 3.19IPDD87 pKa = 3.42NSVIVLPKK95 pKa = 10.21GSFIVSSSITVTDD108 pKa = 3.36ATGITITGHH117 pKa = 6.08GMTEE121 pKa = 4.05TKK123 pKa = 10.54LDD125 pKa = 3.99FSSAPDD131 pKa = 3.59DD132 pKa = 5.17DD133 pKa = 4.18GFKK136 pKa = 10.62FAGGTDD142 pKa = 3.01ITIRR146 pKa = 11.84DD147 pKa = 3.87LGVYY151 pKa = 7.59EE152 pKa = 4.33APKK155 pKa = 10.54NAIKK159 pKa = 10.67ADD161 pKa = 3.81GVNGIHH167 pKa = 6.11ITYY170 pKa = 9.47TSAVWEE176 pKa = 4.25TDD178 pKa = 3.3LEE180 pKa = 4.41EE181 pKa = 5.9GGGNNGAYY189 pKa = 10.13GLYY192 pKa = 9.57PVSSQNVLMEE202 pKa = 4.33NNYY205 pKa = 10.86SKK207 pKa = 11.29GSADD211 pKa = 3.0AGIYY215 pKa = 9.7VGQSNNIVVRR225 pKa = 11.84NNIAEE230 pKa = 4.3HH231 pKa = 5.59NVAGIEE237 pKa = 4.06IEE239 pKa = 4.19NSNKK243 pKa = 9.85ADD245 pKa = 3.39VYY247 pKa = 11.61NNVAFDD253 pKa = 3.64NSAGILSFDD262 pKa = 5.3LPNLPQGYY270 pKa = 9.41GGGVRR275 pKa = 11.84IFNNNTFDD283 pKa = 4.24NNTTNVGAGAVSLAPSGTGILIFATSDD310 pKa = 3.25VEE312 pKa = 4.52IYY314 pKa = 11.3NNTIADD320 pKa = 3.75NDD322 pKa = 4.09TAGIEE327 pKa = 3.91IASYY331 pKa = 10.39FLADD335 pKa = 4.83DD336 pKa = 4.64DD337 pKa = 4.27VANYY341 pKa = 6.68GTNYY345 pKa = 9.1GATMANGWSPLIKK358 pKa = 10.46NINIHH363 pKa = 7.48DD364 pKa = 3.98NTFKK368 pKa = 11.11DD369 pKa = 3.54NSLASPPKK377 pKa = 9.69TGLLEE382 pKa = 5.15DD383 pKa = 5.06IIAGYY388 pKa = 10.28QSAFNSTGSAQTAPAIIYY406 pKa = 9.96GGIGEE411 pKa = 4.74LLSNAGQLTGFDD423 pKa = 3.69ALVGAEE429 pKa = 3.97AKK431 pKa = 10.62ADD433 pKa = 3.68GVDD436 pKa = 3.38YY437 pKa = 10.87DD438 pKa = 4.67AYY440 pKa = 10.81AAGDD444 pKa = 4.54LICSHH449 pKa = 6.88SNINNNNKK457 pKa = 8.19NTLNSNDD464 pKa = 3.75LNVGLVYY471 pKa = 9.6GTDD474 pKa = 3.45ATDD477 pKa = 3.47TNNWNAAGTAPEE489 pKa = 3.81ATLRR493 pKa = 11.84IDD495 pKa = 5.15LIEE498 pKa = 4.61NNTLLDD504 pKa = 3.89CTLEE508 pKa = 4.12RR509 pKa = 11.84LPPAVVTIKK518 pKa = 9.46GTKK521 pKa = 8.95YY522 pKa = 10.7GCTADD527 pKa = 4.35DD528 pKa = 4.99LNLAACSLL536 pKa = 4.0
MM1 pKa = 7.4KK2 pKa = 10.49SSTPSMKK9 pKa = 10.48LGLASAIAVAALGMTGCGGDD29 pKa = 4.21DD30 pKa = 3.37SSSSSKK36 pKa = 9.55STSNLIDD43 pKa = 3.53PCLNADD49 pKa = 3.78CTNSGFILPSNAIYY63 pKa = 10.78VSADD67 pKa = 3.19AADD70 pKa = 4.07GTDD73 pKa = 4.27LKK75 pKa = 11.15NALITALTDD84 pKa = 3.19IPDD87 pKa = 3.42NSVIVLPKK95 pKa = 10.21GSFIVSSSITVTDD108 pKa = 3.36ATGITITGHH117 pKa = 6.08GMTEE121 pKa = 4.05TKK123 pKa = 10.54LDD125 pKa = 3.99FSSAPDD131 pKa = 3.59DD132 pKa = 5.17DD133 pKa = 4.18GFKK136 pKa = 10.62FAGGTDD142 pKa = 3.01ITIRR146 pKa = 11.84DD147 pKa = 3.87LGVYY151 pKa = 7.59EE152 pKa = 4.33APKK155 pKa = 10.54NAIKK159 pKa = 10.67ADD161 pKa = 3.81GVNGIHH167 pKa = 6.11ITYY170 pKa = 9.47TSAVWEE176 pKa = 4.25TDD178 pKa = 3.3LEE180 pKa = 4.41EE181 pKa = 5.9GGGNNGAYY189 pKa = 10.13GLYY192 pKa = 9.57PVSSQNVLMEE202 pKa = 4.33NNYY205 pKa = 10.86SKK207 pKa = 11.29GSADD211 pKa = 3.0AGIYY215 pKa = 9.7VGQSNNIVVRR225 pKa = 11.84NNIAEE230 pKa = 4.3HH231 pKa = 5.59NVAGIEE237 pKa = 4.06IEE239 pKa = 4.19NSNKK243 pKa = 9.85ADD245 pKa = 3.39VYY247 pKa = 11.61NNVAFDD253 pKa = 3.64NSAGILSFDD262 pKa = 5.3LPNLPQGYY270 pKa = 9.41GGGVRR275 pKa = 11.84IFNNNTFDD283 pKa = 4.24NNTTNVGAGAVSLAPSGTGILIFATSDD310 pKa = 3.25VEE312 pKa = 4.52IYY314 pKa = 11.3NNTIADD320 pKa = 3.75NDD322 pKa = 4.09TAGIEE327 pKa = 3.91IASYY331 pKa = 10.39FLADD335 pKa = 4.83DD336 pKa = 4.64DD337 pKa = 4.27VANYY341 pKa = 6.68GTNYY345 pKa = 9.1GATMANGWSPLIKK358 pKa = 10.46NINIHH363 pKa = 7.48DD364 pKa = 3.98NTFKK368 pKa = 11.11DD369 pKa = 3.54NSLASPPKK377 pKa = 9.69TGLLEE382 pKa = 5.15DD383 pKa = 5.06IIAGYY388 pKa = 10.28QSAFNSTGSAQTAPAIIYY406 pKa = 9.96GGIGEE411 pKa = 4.74LLSNAGQLTGFDD423 pKa = 3.69ALVGAEE429 pKa = 3.97AKK431 pKa = 10.62ADD433 pKa = 3.68GVDD436 pKa = 3.38YY437 pKa = 10.87DD438 pKa = 4.67AYY440 pKa = 10.81AAGDD444 pKa = 4.54LICSHH449 pKa = 6.88SNINNNNKK457 pKa = 8.19NTLNSNDD464 pKa = 3.75LNVGLVYY471 pKa = 9.6GTDD474 pKa = 3.45ATDD477 pKa = 3.47TNNWNAAGTAPEE489 pKa = 3.81ATLRR493 pKa = 11.84IDD495 pKa = 5.15LIEE498 pKa = 4.61NNTLLDD504 pKa = 3.89CTLEE508 pKa = 4.12RR509 pKa = 11.84LPPAVVTIKK518 pKa = 9.46GTKK521 pKa = 8.95YY522 pKa = 10.7GCTADD527 pKa = 4.35DD528 pKa = 4.99LNLAACSLL536 pKa = 4.0
Molecular weight: 55.42 kDa
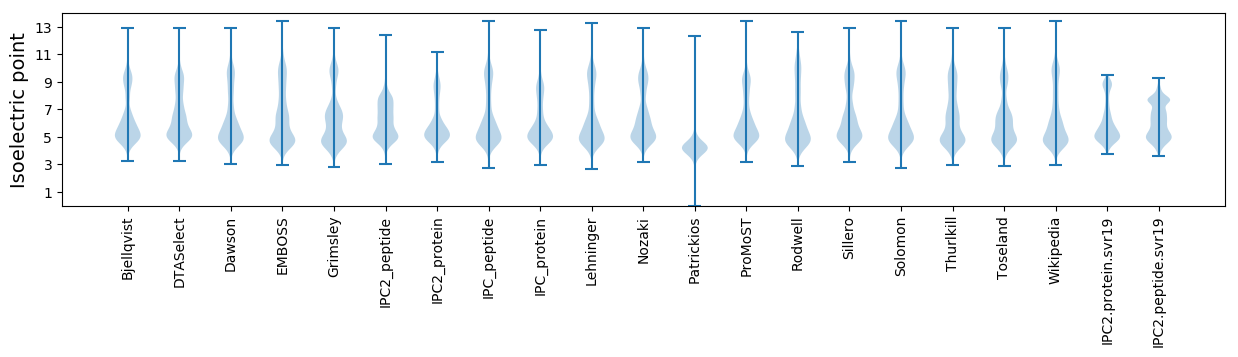
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R4YSX9|R4YSX9_OLEAN Uncharacterized protein OS=Oleispira antarctica RB-8 OX=698738 GN=OLEAN_C11680 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12SGRR28 pKa = 11.84QIISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.59GRR39 pKa = 11.84TKK41 pKa = 10.86LSAA44 pKa = 3.61
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.12SGRR28 pKa = 11.84QIISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.59GRR39 pKa = 11.84TKK41 pKa = 10.86LSAA44 pKa = 3.61
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1295253 |
38 |
4838 |
332.5 |
36.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.508 ± 0.047 | 0.978 ± 0.013 |
5.783 ± 0.039 | 6.326 ± 0.035 |
4.144 ± 0.028 | 6.678 ± 0.043 |
2.189 ± 0.02 | 6.86 ± 0.033 |
5.563 ± 0.036 | 10.4 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.544 ± 0.021 | 4.578 ± 0.037 |
3.736 ± 0.029 | 4.571 ± 0.033 |
4.395 ± 0.031 | 6.882 ± 0.04 |
5.251 ± 0.038 | 6.387 ± 0.035 |
1.243 ± 0.016 | 2.985 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
