
Changjiang tombus-like virus 20
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
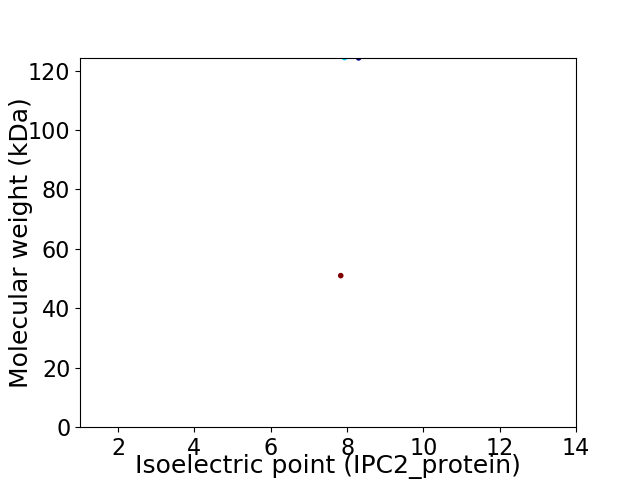
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
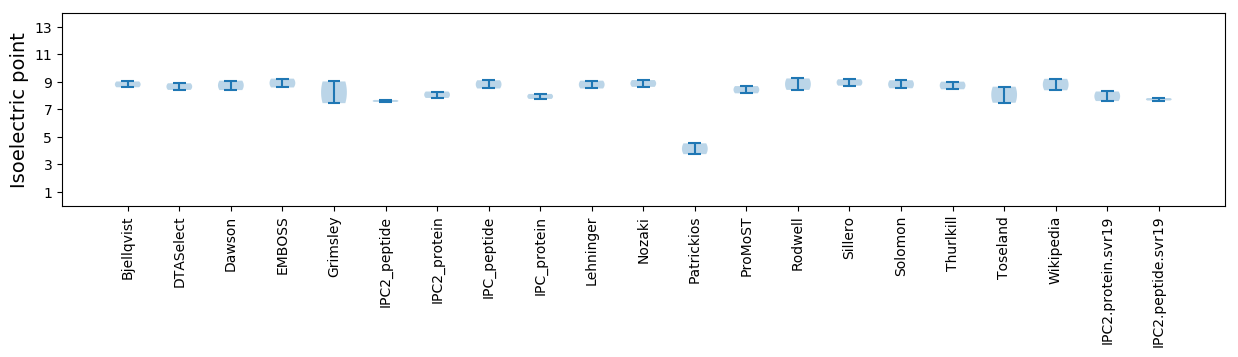
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFX6|A0A1L3KFX6_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 20 OX=1922814 PE=4 SV=1
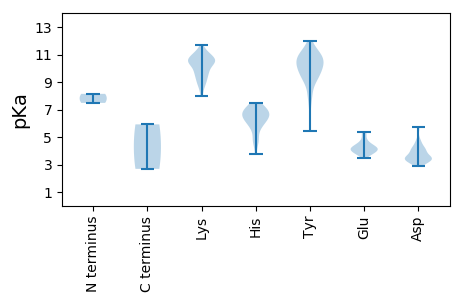
MM1 pKa = 8.14RR2 pKa = 11.84YY3 pKa = 9.31HH4 pKa = 7.3GNYY7 pKa = 9.68CGPSWSDD14 pKa = 3.0GKK16 pKa = 10.27QQLSVAAGDD25 pKa = 3.7QPAIDD30 pKa = 5.58DD31 pKa = 3.89FDD33 pKa = 4.05KK34 pKa = 11.01TCEE37 pKa = 3.94IHH39 pKa = 7.51DD40 pKa = 4.17KK41 pKa = 11.17NYY43 pKa = 11.12ALNTNLYY50 pKa = 9.11EE51 pKa = 4.81ADD53 pKa = 3.17NSFYY57 pKa = 10.16MSNYY61 pKa = 9.78GMGTKK66 pKa = 10.22RR67 pKa = 11.84SLAALAVKK75 pKa = 10.29ANSVLRR81 pKa = 11.84EE82 pKa = 4.01YY83 pKa = 11.11VPNNLPSQVTGPRR96 pKa = 11.84SFNMVKK102 pKa = 10.27NAKK105 pKa = 9.7QPTKK109 pKa = 10.83LRR111 pKa = 11.84GAKK114 pKa = 8.99VAPTKK119 pKa = 10.29RR120 pKa = 11.84TVTKK124 pKa = 10.66NSQQLSTVPASYY136 pKa = 10.73GFSLRR141 pKa = 11.84MQEE144 pKa = 4.13PRR146 pKa = 11.84VQRR149 pKa = 11.84TGKK152 pKa = 7.98NTVIVGSDD160 pKa = 3.53YY161 pKa = 10.22ATSVATSISTGTYY174 pKa = 9.54APSGSILLNPSYY186 pKa = 10.13FASAMLGNVARR197 pKa = 11.84TYY199 pKa = 11.07EE200 pKa = 4.03KK201 pKa = 10.76FRR203 pKa = 11.84FTKK206 pKa = 10.57AVIQYY211 pKa = 9.04IPQVPTTTAGQIVMCSTRR229 pKa = 11.84TVKK232 pKa = 10.54EE233 pKa = 3.44PFINGNSPTFLSRR246 pKa = 11.84ALSQGNAVASPLWKK260 pKa = 9.12EE261 pKa = 3.84TTLTVNCSPEE271 pKa = 3.48WSIVDD276 pKa = 4.14GYY278 pKa = 10.79IDD280 pKa = 4.02TDD282 pKa = 4.07LDD284 pKa = 3.81DD285 pKa = 5.64SIQEE289 pKa = 3.99EE290 pKa = 4.45VQIYY294 pKa = 5.46TTCDD298 pKa = 2.97SVLTTGIIVLHH309 pKa = 4.95YY310 pKa = 9.3TIEE313 pKa = 4.5FKK315 pKa = 11.06DD316 pKa = 3.5PLYY319 pKa = 10.75SYY321 pKa = 10.7HH322 pKa = 7.22SNNIPNPIGNGVFDD336 pKa = 4.79AFSDD340 pKa = 3.71NTPINAVGDD349 pKa = 3.95ALILSNSSTTTGTPSGTIFRR369 pKa = 11.84MVFRR373 pKa = 11.84QQRR376 pKa = 11.84STLPTGVPQWNALANTVTQASSSLATFTNQITSIAMNPGTVLYY419 pKa = 10.33GLQATGGVALFATLEE434 pKa = 4.16SAKK437 pKa = 10.44AGASGYY443 pKa = 7.97ITYY446 pKa = 10.87ASATTVGGVYY456 pKa = 9.53QFIISIVQVGPSIAITTQQ474 pKa = 2.68
MM1 pKa = 8.14RR2 pKa = 11.84YY3 pKa = 9.31HH4 pKa = 7.3GNYY7 pKa = 9.68CGPSWSDD14 pKa = 3.0GKK16 pKa = 10.27QQLSVAAGDD25 pKa = 3.7QPAIDD30 pKa = 5.58DD31 pKa = 3.89FDD33 pKa = 4.05KK34 pKa = 11.01TCEE37 pKa = 3.94IHH39 pKa = 7.51DD40 pKa = 4.17KK41 pKa = 11.17NYY43 pKa = 11.12ALNTNLYY50 pKa = 9.11EE51 pKa = 4.81ADD53 pKa = 3.17NSFYY57 pKa = 10.16MSNYY61 pKa = 9.78GMGTKK66 pKa = 10.22RR67 pKa = 11.84SLAALAVKK75 pKa = 10.29ANSVLRR81 pKa = 11.84EE82 pKa = 4.01YY83 pKa = 11.11VPNNLPSQVTGPRR96 pKa = 11.84SFNMVKK102 pKa = 10.27NAKK105 pKa = 9.7QPTKK109 pKa = 10.83LRR111 pKa = 11.84GAKK114 pKa = 8.99VAPTKK119 pKa = 10.29RR120 pKa = 11.84TVTKK124 pKa = 10.66NSQQLSTVPASYY136 pKa = 10.73GFSLRR141 pKa = 11.84MQEE144 pKa = 4.13PRR146 pKa = 11.84VQRR149 pKa = 11.84TGKK152 pKa = 7.98NTVIVGSDD160 pKa = 3.53YY161 pKa = 10.22ATSVATSISTGTYY174 pKa = 9.54APSGSILLNPSYY186 pKa = 10.13FASAMLGNVARR197 pKa = 11.84TYY199 pKa = 11.07EE200 pKa = 4.03KK201 pKa = 10.76FRR203 pKa = 11.84FTKK206 pKa = 10.57AVIQYY211 pKa = 9.04IPQVPTTTAGQIVMCSTRR229 pKa = 11.84TVKK232 pKa = 10.54EE233 pKa = 3.44PFINGNSPTFLSRR246 pKa = 11.84ALSQGNAVASPLWKK260 pKa = 9.12EE261 pKa = 3.84TTLTVNCSPEE271 pKa = 3.48WSIVDD276 pKa = 4.14GYY278 pKa = 10.79IDD280 pKa = 4.02TDD282 pKa = 4.07LDD284 pKa = 3.81DD285 pKa = 5.64SIQEE289 pKa = 3.99EE290 pKa = 4.45VQIYY294 pKa = 5.46TTCDD298 pKa = 2.97SVLTTGIIVLHH309 pKa = 4.95YY310 pKa = 9.3TIEE313 pKa = 4.5FKK315 pKa = 11.06DD316 pKa = 3.5PLYY319 pKa = 10.75SYY321 pKa = 10.7HH322 pKa = 7.22SNNIPNPIGNGVFDD336 pKa = 4.79AFSDD340 pKa = 3.71NTPINAVGDD349 pKa = 3.95ALILSNSSTTTGTPSGTIFRR369 pKa = 11.84MVFRR373 pKa = 11.84QQRR376 pKa = 11.84STLPTGVPQWNALANTVTQASSSLATFTNQITSIAMNPGTVLYY419 pKa = 10.33GLQATGGVALFATLEE434 pKa = 4.16SAKK437 pKa = 10.44AGASGYY443 pKa = 7.97ITYY446 pKa = 10.87ASATTVGGVYY456 pKa = 9.53QFIISIVQVGPSIAITTQQ474 pKa = 2.68
Molecular weight: 51.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFX6|A0A1L3KFX6_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 20 OX=1922814 PE=4 SV=1
MM1 pKa = 7.48PRR3 pKa = 11.84ASQVKK8 pKa = 8.84TSRR11 pKa = 11.84QPGNRR16 pKa = 11.84ASPVTQKK23 pKa = 8.88NTNRR27 pKa = 11.84DD28 pKa = 3.45SCSSSASTTDD38 pKa = 3.24TVSDD42 pKa = 4.25LVTDD46 pKa = 4.31ARR48 pKa = 11.84PGNALGEE55 pKa = 4.41APSNLSKK62 pKa = 10.72LGKK65 pKa = 10.23DD66 pKa = 3.12GGTAGSAAEE75 pKa = 4.11APLVGEE81 pKa = 4.87LEE83 pKa = 4.42HH84 pKa = 7.14KK85 pKa = 10.5RR86 pKa = 11.84GPKK89 pKa = 9.45SPKK92 pKa = 8.73EE93 pKa = 3.83KK94 pKa = 9.64PGNGVRR100 pKa = 11.84VKK102 pKa = 10.77SKK104 pKa = 9.18TLSPQDD110 pKa = 3.39IKK112 pKa = 11.53NKK114 pKa = 10.25SNSTKK119 pKa = 9.49EE120 pKa = 4.26TVLSGARR127 pKa = 11.84RR128 pKa = 11.84KK129 pKa = 9.12AAPGVRR135 pKa = 11.84LLTQNVMVSRR145 pKa = 11.84NIKK148 pKa = 8.13NTHH151 pKa = 5.73APIAGGQDD159 pKa = 3.07AMHH162 pKa = 6.26QGIGHH167 pKa = 6.66EE168 pKa = 4.32GRR170 pKa = 11.84IKK172 pKa = 9.9FTRR175 pKa = 11.84PWVISPEE182 pKa = 3.9LQIILKK188 pKa = 8.75TRR190 pKa = 11.84FPNRR194 pKa = 11.84LVTTEE199 pKa = 3.55GRR201 pKa = 11.84KK202 pKa = 9.6LYY204 pKa = 9.82NHH206 pKa = 6.9PHH208 pKa = 6.6PLSAIEE214 pKa = 3.68RR215 pKa = 11.84AIIEE219 pKa = 4.11EE220 pKa = 4.43EE221 pKa = 3.83IYY223 pKa = 9.04EE224 pKa = 4.23TLKK227 pKa = 11.38NNDD230 pKa = 3.42CNLIKK235 pKa = 10.66DD236 pKa = 3.47IGGNASRR243 pKa = 11.84HH244 pKa = 3.77KK245 pKa = 8.74TARR248 pKa = 11.84RR249 pKa = 11.84TDD251 pKa = 3.26VHH253 pKa = 6.43SCCPILSPKK262 pKa = 10.49DD263 pKa = 3.29VLRR266 pKa = 11.84NNRR269 pKa = 11.84YY270 pKa = 8.5NATMNYY276 pKa = 6.87CTSKK280 pKa = 11.36AEE282 pKa = 4.26DD283 pKa = 4.07CNVVPDD289 pKa = 4.68GYY291 pKa = 10.23MAIHH295 pKa = 6.32SLYY298 pKa = 9.24YY299 pKa = 10.43LEE301 pKa = 5.03PATVLHH307 pKa = 6.69LVHH310 pKa = 6.65QSRR313 pKa = 11.84KK314 pKa = 9.43KK315 pKa = 9.77ILIASLHH322 pKa = 6.65DD323 pKa = 3.93FSQGYY328 pKa = 10.51GMMHH332 pKa = 6.17YY333 pKa = 10.65NGIEE337 pKa = 3.84HH338 pKa = 6.15EE339 pKa = 4.42TRR341 pKa = 11.84FQIVDD346 pKa = 3.51RR347 pKa = 11.84NTVLMTSSGNDD358 pKa = 3.36GGPYY362 pKa = 8.19THH364 pKa = 6.98SPCFWLRR371 pKa = 11.84KK372 pKa = 9.18CYY374 pKa = 10.66YY375 pKa = 10.06SDD377 pKa = 3.58GKK379 pKa = 10.64RR380 pKa = 11.84AIAWGYY386 pKa = 9.82RR387 pKa = 11.84VVGDD391 pKa = 3.29TYY393 pKa = 10.93IYY395 pKa = 10.55TFTTAPLGLDD405 pKa = 3.46TVKK408 pKa = 10.52EE409 pKa = 3.85RR410 pKa = 11.84DD411 pKa = 3.41MDD413 pKa = 4.06LVSTIRR419 pKa = 11.84NTSHH423 pKa = 6.87HH424 pKa = 6.45GEE426 pKa = 3.94IGATNYY432 pKa = 10.36APLMNYY438 pKa = 9.66LHH440 pKa = 7.46LDD442 pKa = 3.55TVVFRR447 pKa = 11.84SYY449 pKa = 11.62GPLVWAQKK457 pKa = 10.02NSRR460 pKa = 11.84QIFIPKK466 pKa = 10.26GLIQRR471 pKa = 11.84VAFNLVGKK479 pKa = 9.17PRR481 pKa = 11.84NRR483 pKa = 11.84DD484 pKa = 3.35TLIMCVRR491 pKa = 11.84DD492 pKa = 3.75TKK494 pKa = 11.15QEE496 pKa = 3.99LNKK499 pKa = 10.48RR500 pKa = 11.84NLEE503 pKa = 4.16LPDD506 pKa = 3.54SVRR509 pKa = 11.84AEE511 pKa = 3.82LAVFVPPLAFLLHH524 pKa = 7.12LEE526 pKa = 5.13DD527 pKa = 4.49EE528 pKa = 4.7VQSFNRR534 pKa = 11.84LVMPTNQKK542 pKa = 10.37LYY544 pKa = 10.38KK545 pKa = 9.63QLNDD549 pKa = 4.5AINLVKK555 pKa = 10.38PYY557 pKa = 10.87GCLFKK562 pKa = 11.17CFGKK566 pKa = 10.57LKK568 pKa = 9.1HH569 pKa = 5.92QEE571 pKa = 3.84SSVVVDD577 pKa = 4.91EE578 pKa = 4.59YY579 pKa = 11.63NSTRR583 pKa = 11.84TSLDD587 pKa = 3.19TVKK590 pKa = 10.91EE591 pKa = 4.13KK592 pKa = 9.77ITTGFAFLSTTAKK605 pKa = 10.74NAMKK609 pKa = 9.95KK610 pKa = 9.34QRR612 pKa = 11.84SGTYY616 pKa = 8.43FKK618 pKa = 11.12VAIEE622 pKa = 4.13EE623 pKa = 4.14PRR625 pKa = 11.84EE626 pKa = 3.8DD627 pKa = 3.87KK628 pKa = 11.36YY629 pKa = 11.95VMKK632 pKa = 10.61QVATTFSGHH641 pKa = 5.95IPIVPTKK648 pKa = 8.54TQSNTEE654 pKa = 3.58LALRR658 pKa = 11.84NRR660 pKa = 11.84VLAEE664 pKa = 3.66VTPPLEE670 pKa = 4.15GHH672 pKa = 6.86WDD674 pKa = 3.32NMADD678 pKa = 4.01RR679 pKa = 11.84FFNKK683 pKa = 9.49QFLDD687 pKa = 4.13FTQFDD692 pKa = 4.72YY693 pKa = 9.9EE694 pKa = 4.15TDD696 pKa = 3.33EE697 pKa = 5.33DD698 pKa = 4.19KK699 pKa = 11.66AFVKK703 pKa = 9.8WNSGFQPGRR712 pKa = 11.84QKK714 pKa = 10.79DD715 pKa = 3.56HH716 pKa = 5.59EE717 pKa = 4.16RR718 pKa = 11.84ARR720 pKa = 11.84INLKK724 pKa = 10.64NEE726 pKa = 4.37GIKK729 pKa = 10.48NKK731 pKa = 10.02HH732 pKa = 5.42YY733 pKa = 10.12YY734 pKa = 9.99RR735 pKa = 11.84KK736 pKa = 8.15TFMKK740 pKa = 10.43VEE742 pKa = 4.14KK743 pKa = 9.79LDD745 pKa = 3.53KK746 pKa = 10.98SSADD750 pKa = 3.22GVEE753 pKa = 4.25DD754 pKa = 4.27FNPRR758 pKa = 11.84AISGTSHH765 pKa = 6.78EE766 pKa = 5.24SNVVMGPFMTQYY778 pKa = 10.97SKK780 pKa = 11.15QLAKK784 pKa = 10.09QWNGEE789 pKa = 3.77GTFYY793 pKa = 10.07YY794 pKa = 10.44TSGATGEE801 pKa = 4.48STGKK805 pKa = 9.37WMYY808 pKa = 11.64DD809 pKa = 3.01NFTDD813 pKa = 3.52GDD815 pKa = 4.63LIVEE819 pKa = 4.4VDD821 pKa = 4.48FSAYY825 pKa = 10.55DD826 pKa = 3.48STQSRR831 pKa = 11.84DD832 pKa = 2.91SYY834 pKa = 11.15LFEE837 pKa = 5.28KK838 pKa = 10.02RR839 pKa = 11.84VLMAAGMDD847 pKa = 3.3KK848 pKa = 10.99HH849 pKa = 6.28EE850 pKa = 4.63AVCEE854 pKa = 4.26VFKK857 pKa = 10.79HH858 pKa = 4.63QSNMRR863 pKa = 11.84GFTGDD868 pKa = 3.34GIEE871 pKa = 4.03YY872 pKa = 9.92KK873 pKa = 10.66LPYY876 pKa = 9.31GRR878 pKa = 11.84NSGDD882 pKa = 3.62PNTSCGNSLLTAATTEE898 pKa = 4.16YY899 pKa = 11.28VLTKK903 pKa = 10.58VFGPGDD909 pKa = 3.67YY910 pKa = 9.98KK911 pKa = 10.77IKK913 pKa = 11.05VMGDD917 pKa = 3.4DD918 pKa = 3.33NTSIVKK924 pKa = 8.5MANANGVEE932 pKa = 3.89MDD934 pKa = 3.88EE935 pKa = 4.85VIEE938 pKa = 3.96LVKK941 pKa = 10.84KK942 pKa = 10.45EE943 pKa = 3.75FLKK946 pKa = 11.0FGFVAKK952 pKa = 10.43VKK954 pKa = 9.79IHH956 pKa = 6.45YY957 pKa = 9.52DD958 pKa = 3.16VASAEE963 pKa = 4.51FCSSAYY969 pKa = 9.11WPATINNHH977 pKa = 4.65HH978 pKa = 6.93TYY980 pKa = 11.25VMGPKK985 pKa = 9.81PGRR988 pKa = 11.84LLPKK992 pKa = 8.88MGYY995 pKa = 9.18VIKK998 pKa = 10.37DD999 pKa = 3.56LSPGEE1004 pKa = 4.06VKK1006 pKa = 10.72GMFEE1010 pKa = 5.33GYY1012 pKa = 10.89ASTCSHH1018 pKa = 6.35IPVLKK1023 pKa = 10.51QYY1025 pKa = 11.09VNHH1028 pKa = 6.92CLKK1031 pKa = 10.72QMEE1034 pKa = 4.63TIKK1037 pKa = 10.22TKK1039 pKa = 10.87NYY1041 pKa = 7.43TDD1043 pKa = 4.11KK1044 pKa = 10.5EE1045 pKa = 4.24AKK1047 pKa = 10.2YY1048 pKa = 10.75KK1049 pKa = 9.44LTNTGGAVDD1058 pKa = 4.22NDD1060 pKa = 3.18QTAAFFLQRR1069 pKa = 11.84YY1070 pKa = 7.87GLSLDD1075 pKa = 3.49MTIDD1079 pKa = 3.4SLKK1082 pKa = 9.75RR1083 pKa = 11.84TLDD1086 pKa = 3.28GSGPDD1091 pKa = 5.75DD1092 pKa = 3.58MVHH1095 pKa = 5.87WPLLEE1100 pKa = 4.32VIRR1103 pKa = 11.84HH1104 pKa = 4.71VDD1106 pKa = 2.88AA1107 pKa = 5.92
MM1 pKa = 7.48PRR3 pKa = 11.84ASQVKK8 pKa = 8.84TSRR11 pKa = 11.84QPGNRR16 pKa = 11.84ASPVTQKK23 pKa = 8.88NTNRR27 pKa = 11.84DD28 pKa = 3.45SCSSSASTTDD38 pKa = 3.24TVSDD42 pKa = 4.25LVTDD46 pKa = 4.31ARR48 pKa = 11.84PGNALGEE55 pKa = 4.41APSNLSKK62 pKa = 10.72LGKK65 pKa = 10.23DD66 pKa = 3.12GGTAGSAAEE75 pKa = 4.11APLVGEE81 pKa = 4.87LEE83 pKa = 4.42HH84 pKa = 7.14KK85 pKa = 10.5RR86 pKa = 11.84GPKK89 pKa = 9.45SPKK92 pKa = 8.73EE93 pKa = 3.83KK94 pKa = 9.64PGNGVRR100 pKa = 11.84VKK102 pKa = 10.77SKK104 pKa = 9.18TLSPQDD110 pKa = 3.39IKK112 pKa = 11.53NKK114 pKa = 10.25SNSTKK119 pKa = 9.49EE120 pKa = 4.26TVLSGARR127 pKa = 11.84RR128 pKa = 11.84KK129 pKa = 9.12AAPGVRR135 pKa = 11.84LLTQNVMVSRR145 pKa = 11.84NIKK148 pKa = 8.13NTHH151 pKa = 5.73APIAGGQDD159 pKa = 3.07AMHH162 pKa = 6.26QGIGHH167 pKa = 6.66EE168 pKa = 4.32GRR170 pKa = 11.84IKK172 pKa = 9.9FTRR175 pKa = 11.84PWVISPEE182 pKa = 3.9LQIILKK188 pKa = 8.75TRR190 pKa = 11.84FPNRR194 pKa = 11.84LVTTEE199 pKa = 3.55GRR201 pKa = 11.84KK202 pKa = 9.6LYY204 pKa = 9.82NHH206 pKa = 6.9PHH208 pKa = 6.6PLSAIEE214 pKa = 3.68RR215 pKa = 11.84AIIEE219 pKa = 4.11EE220 pKa = 4.43EE221 pKa = 3.83IYY223 pKa = 9.04EE224 pKa = 4.23TLKK227 pKa = 11.38NNDD230 pKa = 3.42CNLIKK235 pKa = 10.66DD236 pKa = 3.47IGGNASRR243 pKa = 11.84HH244 pKa = 3.77KK245 pKa = 8.74TARR248 pKa = 11.84RR249 pKa = 11.84TDD251 pKa = 3.26VHH253 pKa = 6.43SCCPILSPKK262 pKa = 10.49DD263 pKa = 3.29VLRR266 pKa = 11.84NNRR269 pKa = 11.84YY270 pKa = 8.5NATMNYY276 pKa = 6.87CTSKK280 pKa = 11.36AEE282 pKa = 4.26DD283 pKa = 4.07CNVVPDD289 pKa = 4.68GYY291 pKa = 10.23MAIHH295 pKa = 6.32SLYY298 pKa = 9.24YY299 pKa = 10.43LEE301 pKa = 5.03PATVLHH307 pKa = 6.69LVHH310 pKa = 6.65QSRR313 pKa = 11.84KK314 pKa = 9.43KK315 pKa = 9.77ILIASLHH322 pKa = 6.65DD323 pKa = 3.93FSQGYY328 pKa = 10.51GMMHH332 pKa = 6.17YY333 pKa = 10.65NGIEE337 pKa = 3.84HH338 pKa = 6.15EE339 pKa = 4.42TRR341 pKa = 11.84FQIVDD346 pKa = 3.51RR347 pKa = 11.84NTVLMTSSGNDD358 pKa = 3.36GGPYY362 pKa = 8.19THH364 pKa = 6.98SPCFWLRR371 pKa = 11.84KK372 pKa = 9.18CYY374 pKa = 10.66YY375 pKa = 10.06SDD377 pKa = 3.58GKK379 pKa = 10.64RR380 pKa = 11.84AIAWGYY386 pKa = 9.82RR387 pKa = 11.84VVGDD391 pKa = 3.29TYY393 pKa = 10.93IYY395 pKa = 10.55TFTTAPLGLDD405 pKa = 3.46TVKK408 pKa = 10.52EE409 pKa = 3.85RR410 pKa = 11.84DD411 pKa = 3.41MDD413 pKa = 4.06LVSTIRR419 pKa = 11.84NTSHH423 pKa = 6.87HH424 pKa = 6.45GEE426 pKa = 3.94IGATNYY432 pKa = 10.36APLMNYY438 pKa = 9.66LHH440 pKa = 7.46LDD442 pKa = 3.55TVVFRR447 pKa = 11.84SYY449 pKa = 11.62GPLVWAQKK457 pKa = 10.02NSRR460 pKa = 11.84QIFIPKK466 pKa = 10.26GLIQRR471 pKa = 11.84VAFNLVGKK479 pKa = 9.17PRR481 pKa = 11.84NRR483 pKa = 11.84DD484 pKa = 3.35TLIMCVRR491 pKa = 11.84DD492 pKa = 3.75TKK494 pKa = 11.15QEE496 pKa = 3.99LNKK499 pKa = 10.48RR500 pKa = 11.84NLEE503 pKa = 4.16LPDD506 pKa = 3.54SVRR509 pKa = 11.84AEE511 pKa = 3.82LAVFVPPLAFLLHH524 pKa = 7.12LEE526 pKa = 5.13DD527 pKa = 4.49EE528 pKa = 4.7VQSFNRR534 pKa = 11.84LVMPTNQKK542 pKa = 10.37LYY544 pKa = 10.38KK545 pKa = 9.63QLNDD549 pKa = 4.5AINLVKK555 pKa = 10.38PYY557 pKa = 10.87GCLFKK562 pKa = 11.17CFGKK566 pKa = 10.57LKK568 pKa = 9.1HH569 pKa = 5.92QEE571 pKa = 3.84SSVVVDD577 pKa = 4.91EE578 pKa = 4.59YY579 pKa = 11.63NSTRR583 pKa = 11.84TSLDD587 pKa = 3.19TVKK590 pKa = 10.91EE591 pKa = 4.13KK592 pKa = 9.77ITTGFAFLSTTAKK605 pKa = 10.74NAMKK609 pKa = 9.95KK610 pKa = 9.34QRR612 pKa = 11.84SGTYY616 pKa = 8.43FKK618 pKa = 11.12VAIEE622 pKa = 4.13EE623 pKa = 4.14PRR625 pKa = 11.84EE626 pKa = 3.8DD627 pKa = 3.87KK628 pKa = 11.36YY629 pKa = 11.95VMKK632 pKa = 10.61QVATTFSGHH641 pKa = 5.95IPIVPTKK648 pKa = 8.54TQSNTEE654 pKa = 3.58LALRR658 pKa = 11.84NRR660 pKa = 11.84VLAEE664 pKa = 3.66VTPPLEE670 pKa = 4.15GHH672 pKa = 6.86WDD674 pKa = 3.32NMADD678 pKa = 4.01RR679 pKa = 11.84FFNKK683 pKa = 9.49QFLDD687 pKa = 4.13FTQFDD692 pKa = 4.72YY693 pKa = 9.9EE694 pKa = 4.15TDD696 pKa = 3.33EE697 pKa = 5.33DD698 pKa = 4.19KK699 pKa = 11.66AFVKK703 pKa = 9.8WNSGFQPGRR712 pKa = 11.84QKK714 pKa = 10.79DD715 pKa = 3.56HH716 pKa = 5.59EE717 pKa = 4.16RR718 pKa = 11.84ARR720 pKa = 11.84INLKK724 pKa = 10.64NEE726 pKa = 4.37GIKK729 pKa = 10.48NKK731 pKa = 10.02HH732 pKa = 5.42YY733 pKa = 10.12YY734 pKa = 9.99RR735 pKa = 11.84KK736 pKa = 8.15TFMKK740 pKa = 10.43VEE742 pKa = 4.14KK743 pKa = 9.79LDD745 pKa = 3.53KK746 pKa = 10.98SSADD750 pKa = 3.22GVEE753 pKa = 4.25DD754 pKa = 4.27FNPRR758 pKa = 11.84AISGTSHH765 pKa = 6.78EE766 pKa = 5.24SNVVMGPFMTQYY778 pKa = 10.97SKK780 pKa = 11.15QLAKK784 pKa = 10.09QWNGEE789 pKa = 3.77GTFYY793 pKa = 10.07YY794 pKa = 10.44TSGATGEE801 pKa = 4.48STGKK805 pKa = 9.37WMYY808 pKa = 11.64DD809 pKa = 3.01NFTDD813 pKa = 3.52GDD815 pKa = 4.63LIVEE819 pKa = 4.4VDD821 pKa = 4.48FSAYY825 pKa = 10.55DD826 pKa = 3.48STQSRR831 pKa = 11.84DD832 pKa = 2.91SYY834 pKa = 11.15LFEE837 pKa = 5.28KK838 pKa = 10.02RR839 pKa = 11.84VLMAAGMDD847 pKa = 3.3KK848 pKa = 10.99HH849 pKa = 6.28EE850 pKa = 4.63AVCEE854 pKa = 4.26VFKK857 pKa = 10.79HH858 pKa = 4.63QSNMRR863 pKa = 11.84GFTGDD868 pKa = 3.34GIEE871 pKa = 4.03YY872 pKa = 9.92KK873 pKa = 10.66LPYY876 pKa = 9.31GRR878 pKa = 11.84NSGDD882 pKa = 3.62PNTSCGNSLLTAATTEE898 pKa = 4.16YY899 pKa = 11.28VLTKK903 pKa = 10.58VFGPGDD909 pKa = 3.67YY910 pKa = 9.98KK911 pKa = 10.77IKK913 pKa = 11.05VMGDD917 pKa = 3.4DD918 pKa = 3.33NTSIVKK924 pKa = 8.5MANANGVEE932 pKa = 3.89MDD934 pKa = 3.88EE935 pKa = 4.85VIEE938 pKa = 3.96LVKK941 pKa = 10.84KK942 pKa = 10.45EE943 pKa = 3.75FLKK946 pKa = 11.0FGFVAKK952 pKa = 10.43VKK954 pKa = 9.79IHH956 pKa = 6.45YY957 pKa = 9.52DD958 pKa = 3.16VASAEE963 pKa = 4.51FCSSAYY969 pKa = 9.11WPATINNHH977 pKa = 4.65HH978 pKa = 6.93TYY980 pKa = 11.25VMGPKK985 pKa = 9.81PGRR988 pKa = 11.84LLPKK992 pKa = 8.88MGYY995 pKa = 9.18VIKK998 pKa = 10.37DD999 pKa = 3.56LSPGEE1004 pKa = 4.06VKK1006 pKa = 10.72GMFEE1010 pKa = 5.33GYY1012 pKa = 10.89ASTCSHH1018 pKa = 6.35IPVLKK1023 pKa = 10.51QYY1025 pKa = 11.09VNHH1028 pKa = 6.92CLKK1031 pKa = 10.72QMEE1034 pKa = 4.63TIKK1037 pKa = 10.22TKK1039 pKa = 10.87NYY1041 pKa = 7.43TDD1043 pKa = 4.11KK1044 pKa = 10.5EE1045 pKa = 4.24AKK1047 pKa = 10.2YY1048 pKa = 10.75KK1049 pKa = 9.44LTNTGGAVDD1058 pKa = 4.22NDD1060 pKa = 3.18QTAAFFLQRR1069 pKa = 11.84YY1070 pKa = 7.87GLSLDD1075 pKa = 3.49MTIDD1079 pKa = 3.4SLKK1082 pKa = 9.75RR1083 pKa = 11.84TLDD1086 pKa = 3.28GSGPDD1091 pKa = 5.75DD1092 pKa = 3.58MVHH1095 pKa = 5.87WPLLEE1100 pKa = 4.32VIRR1103 pKa = 11.84HH1104 pKa = 4.71VDD1106 pKa = 2.88AA1107 pKa = 5.92
Molecular weight: 124.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1581 |
474 |
1107 |
790.5 |
87.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.958 ± 1.001 | 1.328 ± 0.162 |
5.187 ± 0.822 | 4.491 ± 1.159 |
3.732 ± 0.086 | 7.084 ± 0.053 |
2.404 ± 0.923 | 4.87 ± 0.738 |
6.641 ± 1.683 | 7.147 ± 0.484 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.53 ± 0.374 | 5.946 ± 0.227 |
4.934 ± 0.326 | 3.732 ± 0.788 |
4.681 ± 0.772 | 7.653 ± 1.339 |
8.539 ± 1.564 | 7.021 ± 0.215 |
0.886 ± 0.025 | 4.238 ± 0.239 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
