
Geodermatophilus sp. DSM 44513
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Geodermatophilus; unclassified Geodermatophilus
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
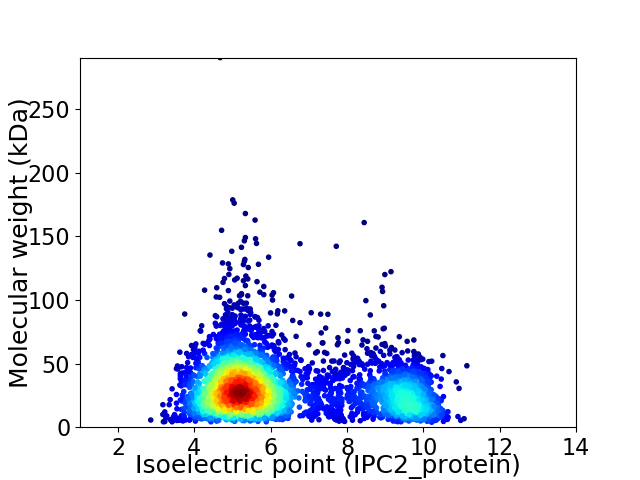
Virtual 2D-PAGE plot for 4421 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5S4ZHC8|A0A5S4ZHC8_9ACTN Osmoprotectant transport system permease protein OS=Geodermatophilus sp. DSM 44513 OX=1528104 GN=JD83_02292 PE=3 SV=1
MM1 pKa = 6.91TRR3 pKa = 11.84SSRR6 pKa = 11.84RR7 pKa = 11.84LTRR10 pKa = 11.84LIATATAVSALAACGGSDD28 pKa = 3.62GGGAGGGGGEE38 pKa = 4.62NEE40 pKa = 4.53ASDD43 pKa = 3.69TGITADD49 pKa = 4.02AIKK52 pKa = 10.41IGAHH56 pKa = 5.39YY57 pKa = 9.82PLTGVAAPGYY67 pKa = 9.3SEE69 pKa = 4.89IPTGVDD75 pKa = 3.32CYY77 pKa = 11.4FDD79 pKa = 3.69FVNAAGGVNGRR90 pKa = 11.84QIDD93 pKa = 3.82YY94 pKa = 10.62VYY96 pKa = 10.95RR97 pKa = 11.84DD98 pKa = 3.69DD99 pKa = 6.02AYY101 pKa = 11.7NPTNTSAVVNEE112 pKa = 4.79LVLQDD117 pKa = 3.99EE118 pKa = 5.2VFMLMGGLGTPTHH131 pKa = 6.06SAVLDD136 pKa = 3.71FLNGEE141 pKa = 4.31GVPDD145 pKa = 4.14LFVASGSLLWDD156 pKa = 3.56NPEE159 pKa = 3.96EE160 pKa = 4.09NPYY163 pKa = 9.44TFGWQPDD170 pKa = 3.75YY171 pKa = 11.01EE172 pKa = 4.39IEE174 pKa = 4.21GKK176 pKa = 10.95VIGQYY181 pKa = 9.67IAEE184 pKa = 4.65NFPEE188 pKa = 4.05ARR190 pKa = 11.84VGLFLQDD197 pKa = 4.18DD198 pKa = 4.27DD199 pKa = 4.79FGEE202 pKa = 4.55DD203 pKa = 3.12GEE205 pKa = 4.67AGVRR209 pKa = 11.84QFIDD213 pKa = 3.6DD214 pKa = 3.89QIVAAEE220 pKa = 4.33RR221 pKa = 11.84YY222 pKa = 9.4VPGNTDD228 pKa = 3.09VGPQLAALQAAGADD242 pKa = 3.51FVVGFNVPSYY252 pKa = 9.64TALSQLTALRR262 pKa = 11.84LGYY265 pKa = 10.24DD266 pKa = 3.55PQWFYY271 pKa = 11.99SNVGSDD277 pKa = 3.53PALVGSLLANFSQGQVSDD295 pKa = 4.04ASLLDD300 pKa = 3.82GALTTEE306 pKa = 4.29YY307 pKa = 10.58LAGVDD312 pKa = 4.0TPDD315 pKa = 5.09DD316 pKa = 3.57PWVQLWQRR324 pKa = 11.84VWDD327 pKa = 3.79EE328 pKa = 4.44CEE330 pKa = 3.93SAEE333 pKa = 5.35GEE335 pKa = 4.13LTNYY339 pKa = 9.37RR340 pKa = 11.84VYY342 pKa = 11.49GMAQAYY348 pKa = 6.17TTIQALQAAGQNPTRR363 pKa = 11.84DD364 pKa = 3.65GLVEE368 pKa = 3.99AVEE371 pKa = 3.91QAGAEE376 pKa = 4.17FEE378 pKa = 4.6GPALAPYY385 pKa = 9.66RR386 pKa = 11.84YY387 pKa = 9.47SAEE390 pKa = 3.64RR391 pKa = 11.84HH392 pKa = 5.55AGVGGVKK399 pKa = 9.77IARR402 pKa = 11.84ITGTSAEE409 pKa = 4.58DD410 pKa = 3.38LTPVLVTDD418 pKa = 4.32NGDD421 pKa = 3.48AAIEE425 pKa = 4.05EE426 pKa = 4.49SDD428 pKa = 4.82AEE430 pKa = 4.31PTTPPEE436 pKa = 3.95SGIPDD441 pKa = 3.72VQPVGG446 pKa = 3.4
MM1 pKa = 6.91TRR3 pKa = 11.84SSRR6 pKa = 11.84RR7 pKa = 11.84LTRR10 pKa = 11.84LIATATAVSALAACGGSDD28 pKa = 3.62GGGAGGGGGEE38 pKa = 4.62NEE40 pKa = 4.53ASDD43 pKa = 3.69TGITADD49 pKa = 4.02AIKK52 pKa = 10.41IGAHH56 pKa = 5.39YY57 pKa = 9.82PLTGVAAPGYY67 pKa = 9.3SEE69 pKa = 4.89IPTGVDD75 pKa = 3.32CYY77 pKa = 11.4FDD79 pKa = 3.69FVNAAGGVNGRR90 pKa = 11.84QIDD93 pKa = 3.82YY94 pKa = 10.62VYY96 pKa = 10.95RR97 pKa = 11.84DD98 pKa = 3.69DD99 pKa = 6.02AYY101 pKa = 11.7NPTNTSAVVNEE112 pKa = 4.79LVLQDD117 pKa = 3.99EE118 pKa = 5.2VFMLMGGLGTPTHH131 pKa = 6.06SAVLDD136 pKa = 3.71FLNGEE141 pKa = 4.31GVPDD145 pKa = 4.14LFVASGSLLWDD156 pKa = 3.56NPEE159 pKa = 3.96EE160 pKa = 4.09NPYY163 pKa = 9.44TFGWQPDD170 pKa = 3.75YY171 pKa = 11.01EE172 pKa = 4.39IEE174 pKa = 4.21GKK176 pKa = 10.95VIGQYY181 pKa = 9.67IAEE184 pKa = 4.65NFPEE188 pKa = 4.05ARR190 pKa = 11.84VGLFLQDD197 pKa = 4.18DD198 pKa = 4.27DD199 pKa = 4.79FGEE202 pKa = 4.55DD203 pKa = 3.12GEE205 pKa = 4.67AGVRR209 pKa = 11.84QFIDD213 pKa = 3.6DD214 pKa = 3.89QIVAAEE220 pKa = 4.33RR221 pKa = 11.84YY222 pKa = 9.4VPGNTDD228 pKa = 3.09VGPQLAALQAAGADD242 pKa = 3.51FVVGFNVPSYY252 pKa = 9.64TALSQLTALRR262 pKa = 11.84LGYY265 pKa = 10.24DD266 pKa = 3.55PQWFYY271 pKa = 11.99SNVGSDD277 pKa = 3.53PALVGSLLANFSQGQVSDD295 pKa = 4.04ASLLDD300 pKa = 3.82GALTTEE306 pKa = 4.29YY307 pKa = 10.58LAGVDD312 pKa = 4.0TPDD315 pKa = 5.09DD316 pKa = 3.57PWVQLWQRR324 pKa = 11.84VWDD327 pKa = 3.79EE328 pKa = 4.44CEE330 pKa = 3.93SAEE333 pKa = 5.35GEE335 pKa = 4.13LTNYY339 pKa = 9.37RR340 pKa = 11.84VYY342 pKa = 11.49GMAQAYY348 pKa = 6.17TTIQALQAAGQNPTRR363 pKa = 11.84DD364 pKa = 3.65GLVEE368 pKa = 3.99AVEE371 pKa = 3.91QAGAEE376 pKa = 4.17FEE378 pKa = 4.6GPALAPYY385 pKa = 9.66RR386 pKa = 11.84YY387 pKa = 9.47SAEE390 pKa = 3.64RR391 pKa = 11.84HH392 pKa = 5.55AGVGGVKK399 pKa = 9.77IARR402 pKa = 11.84ITGTSAEE409 pKa = 4.58DD410 pKa = 3.38LTPVLVTDD418 pKa = 4.32NGDD421 pKa = 3.48AAIEE425 pKa = 4.05EE426 pKa = 4.49SDD428 pKa = 4.82AEE430 pKa = 4.31PTTPPEE436 pKa = 3.95SGIPDD441 pKa = 3.72VQPVGG446 pKa = 3.4
Molecular weight: 46.92 kDa
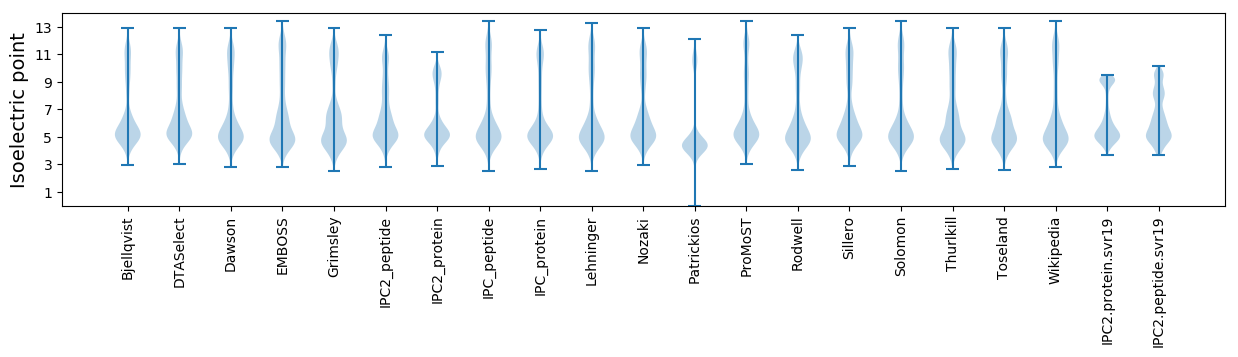
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5S4ZIL3|A0A5S4ZIL3_9ACTN Uncharacterized protein OS=Geodermatophilus sp. DSM 44513 OX=1528104 GN=JD83_02963 PE=4 SV=1
MM1 pKa = 7.35GPAGRR6 pKa = 11.84GRR8 pKa = 11.84SAGRR12 pKa = 11.84AAGDD16 pKa = 3.32RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84GARR22 pKa = 11.84AAGRR26 pKa = 11.84GPAARR31 pKa = 11.84PGRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84HH37 pKa = 5.52HH38 pKa = 5.99GRR40 pKa = 11.84RR41 pKa = 11.84PGARR45 pKa = 11.84RR46 pKa = 11.84GLPPRR51 pKa = 11.84RR52 pKa = 11.84AGPPDD57 pKa = 3.02GRR59 pKa = 11.84LRR61 pKa = 11.84GQRR64 pKa = 11.84GDD66 pKa = 3.35PAGRR70 pKa = 11.84RR71 pKa = 11.84PRR73 pKa = 11.84AAAPPGGRR81 pKa = 11.84PAGAGAGRR89 pKa = 11.84GRR91 pKa = 11.84GAGRR95 pKa = 11.84LRR97 pKa = 11.84GAGPAHH103 pKa = 6.76RR104 pKa = 11.84QRR106 pKa = 11.84GAGGGHH112 pKa = 6.42GCGHH116 pKa = 7.42AAGTGHH122 pKa = 6.99RR123 pKa = 11.84PVAGRR128 pKa = 11.84GAGPGRR134 pKa = 11.84GRR136 pKa = 11.84GRR138 pKa = 11.84PAARR142 pKa = 11.84RR143 pKa = 11.84PAPGARR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84LRR153 pKa = 11.84ALRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84HH159 pKa = 5.38RR160 pKa = 11.84RR161 pKa = 11.84HH162 pKa = 5.77RR163 pKa = 11.84AAGAGLVPAAASPRR177 pKa = 11.84LAAAAGRR184 pKa = 11.84CRR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84RR190 pKa = 11.84GRR192 pKa = 11.84RR193 pKa = 11.84PGHH196 pKa = 4.57RR197 pKa = 11.84TARR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 11.84ALRR205 pKa = 11.84AGQPGVAAGEE215 pKa = 3.95PAGRR219 pKa = 11.84ARR221 pKa = 11.84RR222 pKa = 11.84GAGHH226 pKa = 6.94RR227 pKa = 11.84AGAARR232 pKa = 11.84HH233 pKa = 5.86AGRR236 pKa = 11.84AGGTPAGPGPGVAGRR251 pKa = 11.84LAGALAGRR259 pKa = 11.84GGAGRR264 pKa = 11.84GRR266 pKa = 11.84AARR269 pKa = 11.84RR270 pKa = 11.84GHH272 pKa = 6.35RR273 pKa = 11.84LAGRR277 pKa = 11.84GPRR280 pKa = 11.84RR281 pKa = 11.84GAADD285 pKa = 3.59RR286 pKa = 11.84AGRR289 pKa = 11.84GGRR292 pKa = 11.84LAALAVPPRR301 pKa = 11.84PAAGGGRR308 pKa = 11.84PGRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84GARR316 pKa = 11.84LAAAAGDD323 pKa = 4.0AGLAAAGHH331 pKa = 6.08RR332 pKa = 11.84GGRR335 pKa = 11.84LRR337 pKa = 11.84RR338 pKa = 11.84RR339 pKa = 11.84AGGRR343 pKa = 11.84AGPADD348 pKa = 3.76GAGGGAAGGRR358 pKa = 11.84RR359 pKa = 11.84PGGRR363 pKa = 11.84PGRR366 pKa = 11.84PLPGPAGHH374 pKa = 6.64RR375 pKa = 11.84HH376 pKa = 5.21AAAGAAHH383 pKa = 7.14PPRR386 pKa = 11.84RR387 pKa = 11.84RR388 pKa = 11.84SRR390 pKa = 11.84RR391 pKa = 11.84RR392 pKa = 11.84PGRR395 pKa = 11.84GAVRR399 pKa = 11.84PGRR402 pKa = 11.84RR403 pKa = 11.84GGRR406 pKa = 11.84HH407 pKa = 4.7RR408 pKa = 11.84HH409 pKa = 5.37PLPGRR414 pKa = 11.84GPARR418 pKa = 11.84PAGGRR423 pKa = 11.84GPPGRR428 pKa = 11.84GAAGRR433 pKa = 11.84ARR435 pKa = 11.84AGGAPDD441 pKa = 3.5RR442 pKa = 11.84GQRR445 pKa = 11.84DD446 pKa = 3.34GRR448 pKa = 11.84GARR451 pKa = 11.84PGPGDD456 pKa = 3.23RR457 pKa = 11.84HH458 pKa = 5.35GRR460 pKa = 11.84AAGQRR465 pKa = 11.84PVAGRR470 pKa = 11.84PRR472 pKa = 11.84HH473 pKa = 5.08PARR476 pKa = 11.84RR477 pKa = 11.84PGAVDD482 pKa = 3.35RR483 pKa = 4.97
MM1 pKa = 7.35GPAGRR6 pKa = 11.84GRR8 pKa = 11.84SAGRR12 pKa = 11.84AAGDD16 pKa = 3.32RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84GARR22 pKa = 11.84AAGRR26 pKa = 11.84GPAARR31 pKa = 11.84PGRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84HH37 pKa = 5.52HH38 pKa = 5.99GRR40 pKa = 11.84RR41 pKa = 11.84PGARR45 pKa = 11.84RR46 pKa = 11.84GLPPRR51 pKa = 11.84RR52 pKa = 11.84AGPPDD57 pKa = 3.02GRR59 pKa = 11.84LRR61 pKa = 11.84GQRR64 pKa = 11.84GDD66 pKa = 3.35PAGRR70 pKa = 11.84RR71 pKa = 11.84PRR73 pKa = 11.84AAAPPGGRR81 pKa = 11.84PAGAGAGRR89 pKa = 11.84GRR91 pKa = 11.84GAGRR95 pKa = 11.84LRR97 pKa = 11.84GAGPAHH103 pKa = 6.76RR104 pKa = 11.84QRR106 pKa = 11.84GAGGGHH112 pKa = 6.42GCGHH116 pKa = 7.42AAGTGHH122 pKa = 6.99RR123 pKa = 11.84PVAGRR128 pKa = 11.84GAGPGRR134 pKa = 11.84GRR136 pKa = 11.84GRR138 pKa = 11.84PAARR142 pKa = 11.84RR143 pKa = 11.84PAPGARR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84LRR153 pKa = 11.84ALRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84HH159 pKa = 5.38RR160 pKa = 11.84RR161 pKa = 11.84HH162 pKa = 5.77RR163 pKa = 11.84AAGAGLVPAAASPRR177 pKa = 11.84LAAAAGRR184 pKa = 11.84CRR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84RR190 pKa = 11.84GRR192 pKa = 11.84RR193 pKa = 11.84PGHH196 pKa = 4.57RR197 pKa = 11.84TARR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 11.84ALRR205 pKa = 11.84AGQPGVAAGEE215 pKa = 3.95PAGRR219 pKa = 11.84ARR221 pKa = 11.84RR222 pKa = 11.84GAGHH226 pKa = 6.94RR227 pKa = 11.84AGAARR232 pKa = 11.84HH233 pKa = 5.86AGRR236 pKa = 11.84AGGTPAGPGPGVAGRR251 pKa = 11.84LAGALAGRR259 pKa = 11.84GGAGRR264 pKa = 11.84GRR266 pKa = 11.84AARR269 pKa = 11.84RR270 pKa = 11.84GHH272 pKa = 6.35RR273 pKa = 11.84LAGRR277 pKa = 11.84GPRR280 pKa = 11.84RR281 pKa = 11.84GAADD285 pKa = 3.59RR286 pKa = 11.84AGRR289 pKa = 11.84GGRR292 pKa = 11.84LAALAVPPRR301 pKa = 11.84PAAGGGRR308 pKa = 11.84PGRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84GARR316 pKa = 11.84LAAAAGDD323 pKa = 4.0AGLAAAGHH331 pKa = 6.08RR332 pKa = 11.84GGRR335 pKa = 11.84LRR337 pKa = 11.84RR338 pKa = 11.84RR339 pKa = 11.84AGGRR343 pKa = 11.84AGPADD348 pKa = 3.76GAGGGAAGGRR358 pKa = 11.84RR359 pKa = 11.84PGGRR363 pKa = 11.84PGRR366 pKa = 11.84PLPGPAGHH374 pKa = 6.64RR375 pKa = 11.84HH376 pKa = 5.21AAAGAAHH383 pKa = 7.14PPRR386 pKa = 11.84RR387 pKa = 11.84RR388 pKa = 11.84SRR390 pKa = 11.84RR391 pKa = 11.84RR392 pKa = 11.84PGRR395 pKa = 11.84GAVRR399 pKa = 11.84PGRR402 pKa = 11.84RR403 pKa = 11.84GGRR406 pKa = 11.84HH407 pKa = 4.7RR408 pKa = 11.84HH409 pKa = 5.37PLPGRR414 pKa = 11.84GPARR418 pKa = 11.84PAGGRR423 pKa = 11.84GPPGRR428 pKa = 11.84GAAGRR433 pKa = 11.84ARR435 pKa = 11.84AGGAPDD441 pKa = 3.5RR442 pKa = 11.84GQRR445 pKa = 11.84DD446 pKa = 3.34GRR448 pKa = 11.84GARR451 pKa = 11.84PGPGDD456 pKa = 3.23RR457 pKa = 11.84HH458 pKa = 5.35GRR460 pKa = 11.84AAGQRR465 pKa = 11.84PVAGRR470 pKa = 11.84PRR472 pKa = 11.84HH473 pKa = 5.08PARR476 pKa = 11.84RR477 pKa = 11.84PGAVDD482 pKa = 3.35RR483 pKa = 4.97
Molecular weight: 48.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1365029 |
40 |
2842 |
308.8 |
32.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.864 ± 0.063 | 0.704 ± 0.01 |
6.169 ± 0.028 | 5.296 ± 0.031 |
2.399 ± 0.021 | 9.951 ± 0.032 |
2.052 ± 0.015 | 2.278 ± 0.025 |
1.099 ± 0.02 | 10.843 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.465 ± 0.014 | 1.277 ± 0.016 |
6.46 ± 0.031 | 2.638 ± 0.021 |
8.531 ± 0.039 | 4.51 ± 0.025 |
6.077 ± 0.029 | 10.247 ± 0.036 |
1.461 ± 0.016 | 1.678 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
