
Alkalibacterium sp. 20
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Carnobacteriaceae; Alkalibacterium; unclassified Alkalibacterium
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
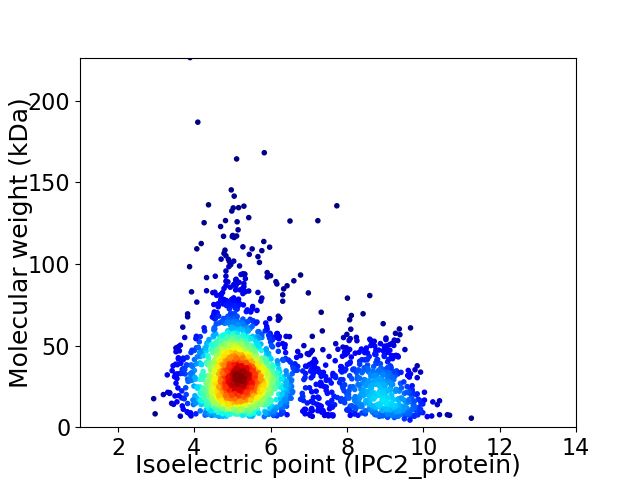
Virtual 2D-PAGE plot for 2261 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
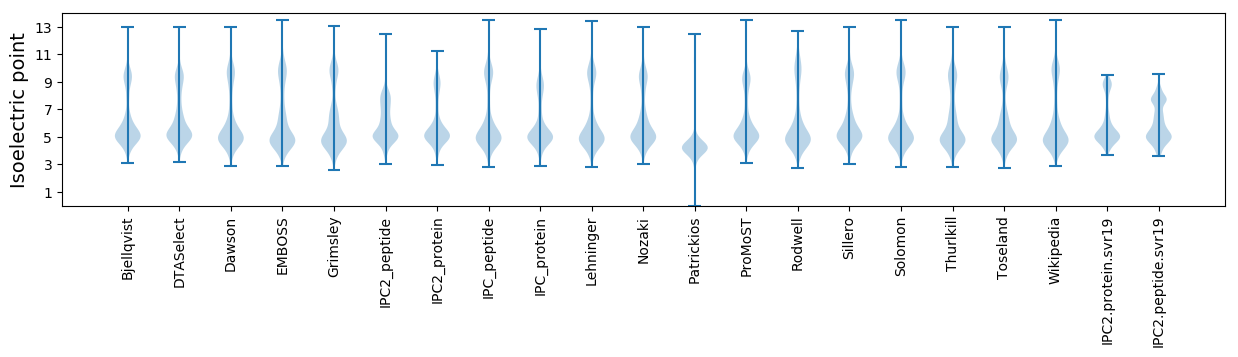
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L8PA90|A0A1L8PA90_9LACT Fe-S cluster assembly ATPase SufC OS=Alkalibacterium sp. 20 OX=1798803 GN=AX762_02805 PE=3 SV=1
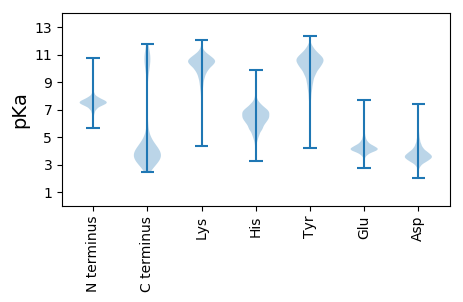
MM1 pKa = 7.53SKK3 pKa = 9.46MKK5 pKa = 10.56SVLKK9 pKa = 10.81YY10 pKa = 8.37GTIVFASSLFLSACGNGDD28 pKa = 4.06VPDD31 pKa = 4.57LDD33 pKa = 4.41EE34 pKa = 5.25DD35 pKa = 4.12ASSEE39 pKa = 4.05EE40 pKa = 4.25GAEE43 pKa = 4.21EE44 pKa = 4.13SAEE47 pKa = 4.07SLPEE51 pKa = 3.62VDD53 pKa = 5.51AGDD56 pKa = 4.24GSTLMVGLTNAPDD69 pKa = 3.66SFNPFYY75 pKa = 10.86RR76 pKa = 11.84PGVAGTWIQRR86 pKa = 11.84FFYY89 pKa = 10.69DD90 pKa = 3.73SLLVMPTADD99 pKa = 3.21SFEE102 pKa = 4.84PGLASFEE109 pKa = 4.46TEE111 pKa = 3.92DD112 pKa = 3.55NQVFTVNIDD121 pKa = 3.43PDD123 pKa = 4.46ANWSDD128 pKa = 4.19GEE130 pKa = 4.33PVSADD135 pKa = 2.97DD136 pKa = 5.14VAFTLNTIADD146 pKa = 4.08PQVEE150 pKa = 4.69TTLGTSVAMIEE161 pKa = 4.49GTDD164 pKa = 3.38SSGVRR169 pKa = 11.84EE170 pKa = 4.3EE171 pKa = 4.36GTEE174 pKa = 3.83EE175 pKa = 3.84LAGVEE180 pKa = 4.27VVDD183 pKa = 4.86EE184 pKa = 4.28KK185 pKa = 10.91TLTITTKK192 pKa = 10.8SPVDD196 pKa = 3.45LAYY199 pKa = 10.79LSEE202 pKa = 3.99FLGFNVLIAPKK213 pKa = 10.17HH214 pKa = 4.8VFEE217 pKa = 6.34DD218 pKa = 3.73IEE220 pKa = 4.45IVDD223 pKa = 3.84IPNSQEE229 pKa = 3.28ATMPSVFSGAYY240 pKa = 9.31QFVEE244 pKa = 4.27YY245 pKa = 11.07DD246 pKa = 3.0NDD248 pKa = 3.85NYY250 pKa = 10.86VHH252 pKa = 7.05LEE254 pKa = 3.8ANPDD258 pKa = 3.69YY259 pKa = 10.89FRR261 pKa = 11.84GAPAIEE267 pKa = 3.91TVYY270 pKa = 10.92ARR272 pKa = 11.84VMNGTSLITEE282 pKa = 4.49FQAGNLHH289 pKa = 5.38MTAGGGIGMVPVQDD303 pKa = 3.42IGLLEE308 pKa = 5.15DD309 pKa = 3.42IEE311 pKa = 4.6GLVVEE316 pKa = 5.03EE317 pKa = 4.64HH318 pKa = 7.04PSFNGQYY325 pKa = 9.62MVINNEE331 pKa = 3.79KK332 pKa = 10.16FDD334 pKa = 4.0NLEE337 pKa = 3.61VRR339 pKa = 11.84QAFAYY344 pKa = 10.12ALNRR348 pKa = 11.84EE349 pKa = 4.32LTVEE353 pKa = 3.95NLLGGRR359 pKa = 11.84GEE361 pKa = 4.44VLASTYY367 pKa = 11.11SSASPYY373 pKa = 10.39KK374 pKa = 10.32DD375 pKa = 4.47DD376 pKa = 3.61SLEE379 pKa = 4.02PLEE382 pKa = 4.76YY383 pKa = 10.66DD384 pKa = 3.4PEE386 pKa = 4.3LAKK389 pKa = 10.93EE390 pKa = 4.05MLEE393 pKa = 4.02EE394 pKa = 4.94AGFDD398 pKa = 3.4FDD400 pKa = 4.11TPLEE404 pKa = 4.24FVVPTGNAVRR414 pKa = 11.84EE415 pKa = 4.2QNGNLIEE422 pKa = 4.1QWFSEE427 pKa = 3.8IGVTVNLNSYY437 pKa = 10.2DD438 pKa = 3.69FPTWLSMAQDD448 pKa = 3.96LDD450 pKa = 3.9YY451 pKa = 11.21DD452 pKa = 3.74IGLMGWGHH460 pKa = 5.48TVDD463 pKa = 4.61PNIASYY469 pKa = 10.3IQTGGASNNMAVSDD483 pKa = 4.39PVVDD487 pKa = 4.96DD488 pKa = 5.7LIEE491 pKa = 4.02QGMAGTDD498 pKa = 3.49FDD500 pKa = 4.04EE501 pKa = 5.48RR502 pKa = 11.84YY503 pKa = 9.52PIYY506 pKa = 10.98AEE508 pKa = 4.44LQQHH512 pKa = 5.74MQDD515 pKa = 3.48EE516 pKa = 4.6MQIVPLYY523 pKa = 10.45SDD525 pKa = 3.29SQFSVQVDD533 pKa = 3.6NLNGGIKK540 pKa = 9.76EE541 pKa = 3.97YY542 pKa = 9.94WAGSLHH548 pKa = 7.38DD549 pKa = 3.87LHH551 pKa = 7.01EE552 pKa = 4.38WTLDD556 pKa = 3.47TEE558 pKa = 4.57EE559 pKa = 4.16
MM1 pKa = 7.53SKK3 pKa = 9.46MKK5 pKa = 10.56SVLKK9 pKa = 10.81YY10 pKa = 8.37GTIVFASSLFLSACGNGDD28 pKa = 4.06VPDD31 pKa = 4.57LDD33 pKa = 4.41EE34 pKa = 5.25DD35 pKa = 4.12ASSEE39 pKa = 4.05EE40 pKa = 4.25GAEE43 pKa = 4.21EE44 pKa = 4.13SAEE47 pKa = 4.07SLPEE51 pKa = 3.62VDD53 pKa = 5.51AGDD56 pKa = 4.24GSTLMVGLTNAPDD69 pKa = 3.66SFNPFYY75 pKa = 10.86RR76 pKa = 11.84PGVAGTWIQRR86 pKa = 11.84FFYY89 pKa = 10.69DD90 pKa = 3.73SLLVMPTADD99 pKa = 3.21SFEE102 pKa = 4.84PGLASFEE109 pKa = 4.46TEE111 pKa = 3.92DD112 pKa = 3.55NQVFTVNIDD121 pKa = 3.43PDD123 pKa = 4.46ANWSDD128 pKa = 4.19GEE130 pKa = 4.33PVSADD135 pKa = 2.97DD136 pKa = 5.14VAFTLNTIADD146 pKa = 4.08PQVEE150 pKa = 4.69TTLGTSVAMIEE161 pKa = 4.49GTDD164 pKa = 3.38SSGVRR169 pKa = 11.84EE170 pKa = 4.3EE171 pKa = 4.36GTEE174 pKa = 3.83EE175 pKa = 3.84LAGVEE180 pKa = 4.27VVDD183 pKa = 4.86EE184 pKa = 4.28KK185 pKa = 10.91TLTITTKK192 pKa = 10.8SPVDD196 pKa = 3.45LAYY199 pKa = 10.79LSEE202 pKa = 3.99FLGFNVLIAPKK213 pKa = 10.17HH214 pKa = 4.8VFEE217 pKa = 6.34DD218 pKa = 3.73IEE220 pKa = 4.45IVDD223 pKa = 3.84IPNSQEE229 pKa = 3.28ATMPSVFSGAYY240 pKa = 9.31QFVEE244 pKa = 4.27YY245 pKa = 11.07DD246 pKa = 3.0NDD248 pKa = 3.85NYY250 pKa = 10.86VHH252 pKa = 7.05LEE254 pKa = 3.8ANPDD258 pKa = 3.69YY259 pKa = 10.89FRR261 pKa = 11.84GAPAIEE267 pKa = 3.91TVYY270 pKa = 10.92ARR272 pKa = 11.84VMNGTSLITEE282 pKa = 4.49FQAGNLHH289 pKa = 5.38MTAGGGIGMVPVQDD303 pKa = 3.42IGLLEE308 pKa = 5.15DD309 pKa = 3.42IEE311 pKa = 4.6GLVVEE316 pKa = 5.03EE317 pKa = 4.64HH318 pKa = 7.04PSFNGQYY325 pKa = 9.62MVINNEE331 pKa = 3.79KK332 pKa = 10.16FDD334 pKa = 4.0NLEE337 pKa = 3.61VRR339 pKa = 11.84QAFAYY344 pKa = 10.12ALNRR348 pKa = 11.84EE349 pKa = 4.32LTVEE353 pKa = 3.95NLLGGRR359 pKa = 11.84GEE361 pKa = 4.44VLASTYY367 pKa = 11.11SSASPYY373 pKa = 10.39KK374 pKa = 10.32DD375 pKa = 4.47DD376 pKa = 3.61SLEE379 pKa = 4.02PLEE382 pKa = 4.76YY383 pKa = 10.66DD384 pKa = 3.4PEE386 pKa = 4.3LAKK389 pKa = 10.93EE390 pKa = 4.05MLEE393 pKa = 4.02EE394 pKa = 4.94AGFDD398 pKa = 3.4FDD400 pKa = 4.11TPLEE404 pKa = 4.24FVVPTGNAVRR414 pKa = 11.84EE415 pKa = 4.2QNGNLIEE422 pKa = 4.1QWFSEE427 pKa = 3.8IGVTVNLNSYY437 pKa = 10.2DD438 pKa = 3.69FPTWLSMAQDD448 pKa = 3.96LDD450 pKa = 3.9YY451 pKa = 11.21DD452 pKa = 3.74IGLMGWGHH460 pKa = 5.48TVDD463 pKa = 4.61PNIASYY469 pKa = 10.3IQTGGASNNMAVSDD483 pKa = 4.39PVVDD487 pKa = 4.96DD488 pKa = 5.7LIEE491 pKa = 4.02QGMAGTDD498 pKa = 3.49FDD500 pKa = 4.04EE501 pKa = 5.48RR502 pKa = 11.84YY503 pKa = 9.52PIYY506 pKa = 10.98AEE508 pKa = 4.44LQQHH512 pKa = 5.74MQDD515 pKa = 3.48EE516 pKa = 4.6MQIVPLYY523 pKa = 10.45SDD525 pKa = 3.29SQFSVQVDD533 pKa = 3.6NLNGGIKK540 pKa = 9.76EE541 pKa = 3.97YY542 pKa = 9.94WAGSLHH548 pKa = 7.38DD549 pKa = 3.87LHH551 pKa = 7.01EE552 pKa = 4.38WTLDD556 pKa = 3.47TEE558 pKa = 4.57EE559 pKa = 4.16
Molecular weight: 61.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L8PPM8|A0A1L8PPM8_9LACT Formate acetyltransferase OS=Alkalibacterium sp. 20 OX=1798803 GN=AX762_00280 PE=3 SV=1
MM1 pKa = 7.3AQGLTHH7 pKa = 5.92QPKK10 pKa = 8.82KK11 pKa = 10.23RR12 pKa = 11.84KK13 pKa = 7.68RR14 pKa = 11.84QRR16 pKa = 11.84VHH18 pKa = 6.34GFRR21 pKa = 11.84KK22 pKa = 10.03RR23 pKa = 11.84MSTKK27 pKa = 9.73SGRR30 pKa = 11.84NILRR34 pKa = 11.84NRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 8.93GRR41 pKa = 11.84KK42 pKa = 9.14NISAA46 pKa = 4.08
MM1 pKa = 7.3AQGLTHH7 pKa = 5.92QPKK10 pKa = 8.82KK11 pKa = 10.23RR12 pKa = 11.84KK13 pKa = 7.68RR14 pKa = 11.84QRR16 pKa = 11.84VHH18 pKa = 6.34GFRR21 pKa = 11.84KK22 pKa = 10.03RR23 pKa = 11.84MSTKK27 pKa = 9.73SGRR30 pKa = 11.84NILRR34 pKa = 11.84NRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 8.93GRR41 pKa = 11.84KK42 pKa = 9.14NISAA46 pKa = 4.08
Molecular weight: 5.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
687154 |
37 |
2011 |
303.9 |
34.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.66 ± 0.051 | 0.481 ± 0.011 |
5.889 ± 0.049 | 8.077 ± 0.072 |
4.449 ± 0.043 | 6.517 ± 0.048 |
1.937 ± 0.021 | 7.773 ± 0.05 |
6.864 ± 0.058 | 9.676 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.743 ± 0.022 | 4.697 ± 0.034 |
3.321 ± 0.029 | 3.472 ± 0.032 |
3.934 ± 0.032 | 6.261 ± 0.035 |
5.749 ± 0.029 | 6.927 ± 0.039 |
0.905 ± 0.019 | 3.668 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
