
Aeoliella mucimassa
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales;
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
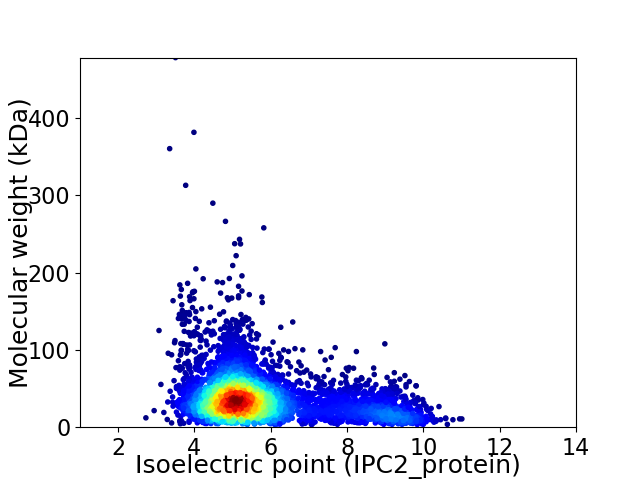
Virtual 2D-PAGE plot for 5127 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
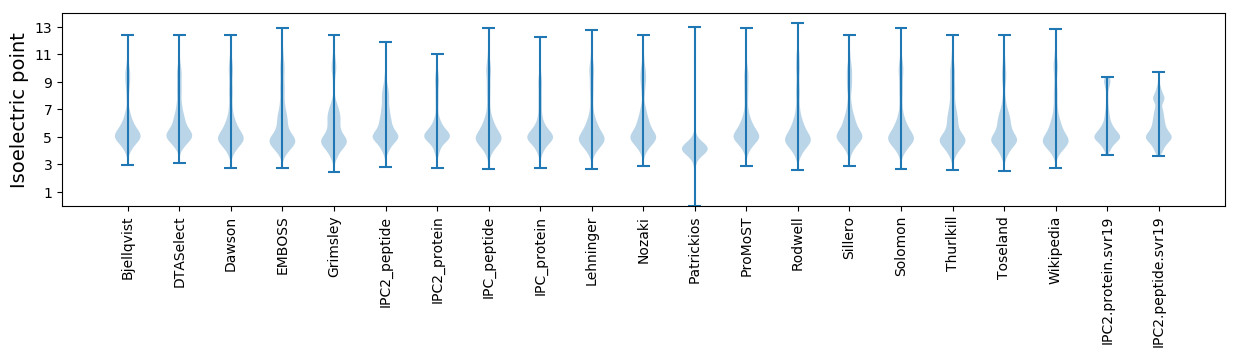
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A518AQP7|A0A518AQP7_9BACT Uncharacterized protein OS=Aeoliella mucimassa OX=2527972 GN=Pan181_32620 PE=4 SV=1
MM1 pKa = 7.69HH2 pKa = 7.21NRR4 pKa = 11.84PLYY7 pKa = 9.37WALLLLSATALNHH20 pKa = 4.61VWLRR24 pKa = 11.84SCTAAPAAFWFSTDD38 pKa = 2.7ATLPGTPGVPEE49 pKa = 3.92FTSAVGIPRR58 pKa = 11.84TLHH61 pKa = 5.84IWAQPNTLTSGDD73 pKa = 3.65WNASTNPFLKK83 pKa = 10.21FQNVSLDD90 pKa = 3.3IVSPEE95 pKa = 3.88STFDD99 pKa = 3.41IDD101 pKa = 3.39PDD103 pKa = 4.27SIVVYY108 pKa = 10.49NPTYY112 pKa = 11.0ASGEE116 pKa = 4.03DD117 pKa = 3.25RR118 pKa = 11.84FSNVGDD124 pKa = 3.64SSTGLTEE131 pKa = 4.23SDD133 pKa = 3.36GSALGSYY140 pKa = 10.83DD141 pKa = 4.03DD142 pKa = 5.16LPIGFSKK149 pKa = 10.94GLLDD153 pKa = 3.75LQGFSMPANYY163 pKa = 10.25AQGFGVTCDD172 pKa = 4.33ASDD175 pKa = 4.17SYY177 pKa = 11.73CGTTDD182 pKa = 2.91SGSPAWLFASFAVTPTSDD200 pKa = 3.11TGSVEE205 pKa = 3.74FWMQIGRR212 pKa = 11.84NGINGVDD219 pKa = 3.82ASDD222 pKa = 3.96ALLGSGTIEE231 pKa = 3.91VQFGIDD237 pKa = 3.42TEE239 pKa = 4.71GFAPFDD245 pKa = 3.65YY246 pKa = 10.77DD247 pKa = 3.94AEE249 pKa = 4.3YY250 pKa = 11.04DD251 pKa = 4.09RR252 pKa = 11.84KK253 pKa = 9.59LTLANDD259 pKa = 4.07DD260 pKa = 3.91SDD262 pKa = 4.17VTLLLAGPGDD272 pKa = 4.16YY273 pKa = 10.94NADD276 pKa = 3.69GAVDD280 pKa = 3.76EE281 pKa = 5.31LDD283 pKa = 3.54YY284 pKa = 11.5AVWKK288 pKa = 10.55KK289 pKa = 11.02DD290 pKa = 3.28FGTNSYY296 pKa = 10.86DD297 pKa = 3.83ADD299 pKa = 4.06GNGDD303 pKa = 4.28GIVSLADD310 pKa = 3.23YY311 pKa = 7.58TIWRR315 pKa = 11.84DD316 pKa = 3.59NLDD319 pKa = 3.63ATSTGALASLNVSQVPEE336 pKa = 4.07PSTFAMLAGFTTLAWNLSSSSRR358 pKa = 11.84ARR360 pKa = 11.84HH361 pKa = 5.64SSNCEE366 pKa = 3.4
MM1 pKa = 7.69HH2 pKa = 7.21NRR4 pKa = 11.84PLYY7 pKa = 9.37WALLLLSATALNHH20 pKa = 4.61VWLRR24 pKa = 11.84SCTAAPAAFWFSTDD38 pKa = 2.7ATLPGTPGVPEE49 pKa = 3.92FTSAVGIPRR58 pKa = 11.84TLHH61 pKa = 5.84IWAQPNTLTSGDD73 pKa = 3.65WNASTNPFLKK83 pKa = 10.21FQNVSLDD90 pKa = 3.3IVSPEE95 pKa = 3.88STFDD99 pKa = 3.41IDD101 pKa = 3.39PDD103 pKa = 4.27SIVVYY108 pKa = 10.49NPTYY112 pKa = 11.0ASGEE116 pKa = 4.03DD117 pKa = 3.25RR118 pKa = 11.84FSNVGDD124 pKa = 3.64SSTGLTEE131 pKa = 4.23SDD133 pKa = 3.36GSALGSYY140 pKa = 10.83DD141 pKa = 4.03DD142 pKa = 5.16LPIGFSKK149 pKa = 10.94GLLDD153 pKa = 3.75LQGFSMPANYY163 pKa = 10.25AQGFGVTCDD172 pKa = 4.33ASDD175 pKa = 4.17SYY177 pKa = 11.73CGTTDD182 pKa = 2.91SGSPAWLFASFAVTPTSDD200 pKa = 3.11TGSVEE205 pKa = 3.74FWMQIGRR212 pKa = 11.84NGINGVDD219 pKa = 3.82ASDD222 pKa = 3.96ALLGSGTIEE231 pKa = 3.91VQFGIDD237 pKa = 3.42TEE239 pKa = 4.71GFAPFDD245 pKa = 3.65YY246 pKa = 10.77DD247 pKa = 3.94AEE249 pKa = 4.3YY250 pKa = 11.04DD251 pKa = 4.09RR252 pKa = 11.84KK253 pKa = 9.59LTLANDD259 pKa = 4.07DD260 pKa = 3.91SDD262 pKa = 4.17VTLLLAGPGDD272 pKa = 4.16YY273 pKa = 10.94NADD276 pKa = 3.69GAVDD280 pKa = 3.76EE281 pKa = 5.31LDD283 pKa = 3.54YY284 pKa = 11.5AVWKK288 pKa = 10.55KK289 pKa = 11.02DD290 pKa = 3.28FGTNSYY296 pKa = 10.86DD297 pKa = 3.83ADD299 pKa = 4.06GNGDD303 pKa = 4.28GIVSLADD310 pKa = 3.23YY311 pKa = 7.58TIWRR315 pKa = 11.84DD316 pKa = 3.59NLDD319 pKa = 3.63ATSTGALASLNVSQVPEE336 pKa = 4.07PSTFAMLAGFTTLAWNLSSSSRR358 pKa = 11.84ARR360 pKa = 11.84HH361 pKa = 5.64SSNCEE366 pKa = 3.4
Molecular weight: 38.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518AKX6|A0A518AKX6_9BACT Dihydroorotase OS=Aeoliella mucimassa OX=2527972 GN=pyrC PE=3 SV=1
MM1 pKa = 7.44MKK3 pKa = 10.19RR4 pKa = 11.84AFDD7 pKa = 3.44IVGALVLLAVLSPLFLLVAVWMKK30 pKa = 10.57YY31 pKa = 6.85EE32 pKa = 3.87SRR34 pKa = 11.84GPVLCRR40 pKa = 11.84EE41 pKa = 4.0RR42 pKa = 11.84RR43 pKa = 11.84AGRR46 pKa = 11.84GGLPFRR52 pKa = 11.84TFKK55 pKa = 10.77FRR57 pKa = 11.84VLHH60 pKa = 6.18ASSAALCGTSTPRR73 pKa = 11.84NDD75 pKa = 3.8PQLSASGRR83 pKa = 11.84LMRR86 pKa = 11.84RR87 pKa = 11.84FRR89 pKa = 11.84IHH91 pKa = 7.0EE92 pKa = 4.4LPQLLNVLRR101 pKa = 11.84GEE103 pKa = 4.2MSLVGPGPEE112 pKa = 3.79LVDD115 pKa = 4.24RR116 pKa = 11.84VDD118 pKa = 3.86SLSGSDD124 pKa = 4.44KK125 pKa = 10.86RR126 pKa = 11.84LLNLRR131 pKa = 11.84PGMTDD136 pKa = 3.17WASLWNSDD144 pKa = 3.98EE145 pKa = 4.33ASSLAGAPDD154 pKa = 3.44RR155 pKa = 11.84EE156 pKa = 4.07LAYY159 pKa = 10.78DD160 pKa = 3.75RR161 pKa = 11.84FIRR164 pKa = 11.84PRR166 pKa = 11.84KK167 pKa = 9.49LRR169 pKa = 11.84LQKK172 pKa = 10.79YY173 pKa = 7.69YY174 pKa = 11.12LEE176 pKa = 4.25NRR178 pKa = 11.84SLGMDD183 pKa = 3.98LKK185 pKa = 10.79ILTFTVLRR193 pKa = 11.84VINRR197 pKa = 11.84QFLPRR202 pKa = 11.84EE203 pKa = 3.87LHH205 pKa = 5.74NFPTFGEE212 pKa = 4.0LRR214 pKa = 11.84AEE216 pKa = 4.31VLCLEE221 pKa = 4.32SSAMIRR227 pKa = 11.84KK228 pKa = 8.49HH229 pKa = 5.67AAA231 pKa = 2.83
MM1 pKa = 7.44MKK3 pKa = 10.19RR4 pKa = 11.84AFDD7 pKa = 3.44IVGALVLLAVLSPLFLLVAVWMKK30 pKa = 10.57YY31 pKa = 6.85EE32 pKa = 3.87SRR34 pKa = 11.84GPVLCRR40 pKa = 11.84EE41 pKa = 4.0RR42 pKa = 11.84RR43 pKa = 11.84AGRR46 pKa = 11.84GGLPFRR52 pKa = 11.84TFKK55 pKa = 10.77FRR57 pKa = 11.84VLHH60 pKa = 6.18ASSAALCGTSTPRR73 pKa = 11.84NDD75 pKa = 3.8PQLSASGRR83 pKa = 11.84LMRR86 pKa = 11.84RR87 pKa = 11.84FRR89 pKa = 11.84IHH91 pKa = 7.0EE92 pKa = 4.4LPQLLNVLRR101 pKa = 11.84GEE103 pKa = 4.2MSLVGPGPEE112 pKa = 3.79LVDD115 pKa = 4.24RR116 pKa = 11.84VDD118 pKa = 3.86SLSGSDD124 pKa = 4.44KK125 pKa = 10.86RR126 pKa = 11.84LLNLRR131 pKa = 11.84PGMTDD136 pKa = 3.17WASLWNSDD144 pKa = 3.98EE145 pKa = 4.33ASSLAGAPDD154 pKa = 3.44RR155 pKa = 11.84EE156 pKa = 4.07LAYY159 pKa = 10.78DD160 pKa = 3.75RR161 pKa = 11.84FIRR164 pKa = 11.84PRR166 pKa = 11.84KK167 pKa = 9.49LRR169 pKa = 11.84LQKK172 pKa = 10.79YY173 pKa = 7.69YY174 pKa = 11.12LEE176 pKa = 4.25NRR178 pKa = 11.84SLGMDD183 pKa = 3.98LKK185 pKa = 10.79ILTFTVLRR193 pKa = 11.84VINRR197 pKa = 11.84QFLPRR202 pKa = 11.84EE203 pKa = 3.87LHH205 pKa = 5.74NFPTFGEE212 pKa = 4.0LRR214 pKa = 11.84AEE216 pKa = 4.31VLCLEE221 pKa = 4.32SSAMIRR227 pKa = 11.84KK228 pKa = 8.49HH229 pKa = 5.67AAA231 pKa = 2.83
Molecular weight: 26.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1840367 |
29 |
4569 |
359.0 |
39.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.891 ± 0.038 | 1.114 ± 0.014 |
6.137 ± 0.03 | 6.469 ± 0.045 |
3.488 ± 0.021 | 7.84 ± 0.047 |
2.158 ± 0.018 | 4.63 ± 0.021 |
3.312 ± 0.03 | 9.913 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.274 ± 0.016 | 3.328 ± 0.027 |
5.101 ± 0.025 | 4.115 ± 0.026 |
5.986 ± 0.04 | 6.527 ± 0.033 |
5.932 ± 0.038 | 7.404 ± 0.031 |
1.608 ± 0.016 | 2.774 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
