
Allium cepa amalgavirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; Amalgavirus
Average proteome isoelectric point is 7.96
Get precalculated fractions of proteins
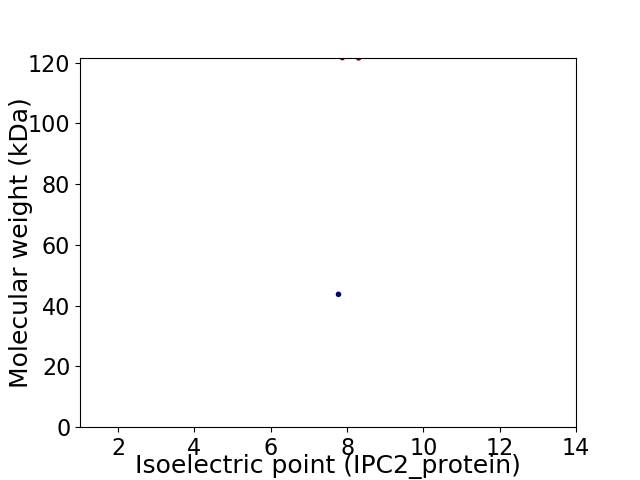
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
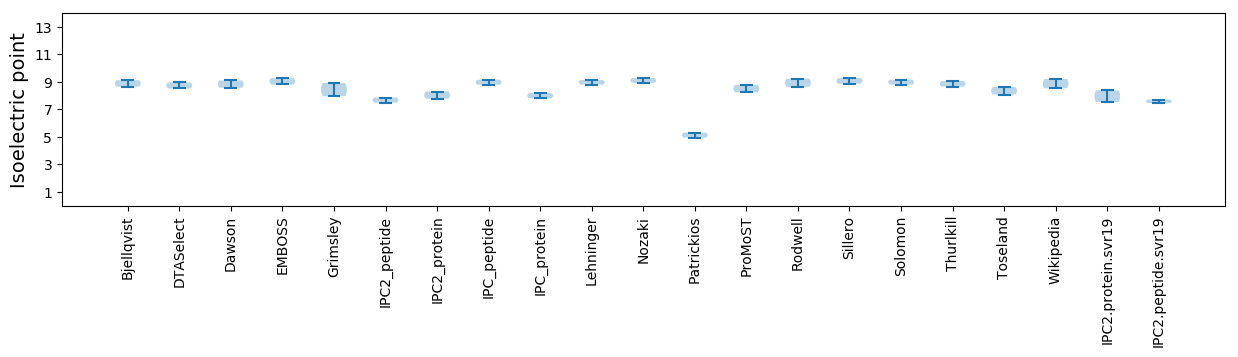
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5MGZ5|A0A2H5MGZ5_9VIRU ORF1+2p OS=Allium cepa amalgavirus 2 OX=2058779 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.62ASSSGPDD8 pKa = 3.11PTQYY12 pKa = 11.62LDD14 pKa = 3.09ITLLSDD20 pKa = 3.88PVAEE24 pKa = 4.38SVSLQDD30 pKa = 4.33AVQHH34 pKa = 6.0LASIGVRR41 pKa = 11.84VEE43 pKa = 3.59RR44 pKa = 11.84FTRR47 pKa = 11.84DD48 pKa = 2.87SIHH51 pKa = 6.77AMRR54 pKa = 11.84MSVPAYY60 pKa = 9.43VKK62 pKa = 10.04EE63 pKa = 3.64IRR65 pKa = 11.84ILGNISDD72 pKa = 4.59RR73 pKa = 11.84EE74 pKa = 4.01LLKK77 pKa = 11.05NVMLQGVKK85 pKa = 10.78NNVLSLPEE93 pKa = 4.53DD94 pKa = 3.56VTPQMVLSFARR105 pKa = 11.84WLKK108 pKa = 9.84RR109 pKa = 11.84GAGARR114 pKa = 11.84VLADD118 pKa = 3.35QQHH121 pKa = 6.27LLKK124 pKa = 10.42INKK127 pKa = 9.06KK128 pKa = 10.14AVGDD132 pKa = 3.92STPDD136 pKa = 2.91IVAFAHH142 pKa = 7.2LLDD145 pKa = 4.1QQVQDD150 pKa = 3.73LTNAKK155 pKa = 9.96KK156 pKa = 10.16RR157 pKa = 11.84VQHH160 pKa = 6.03SSQVRR165 pKa = 11.84IDD167 pKa = 3.75EE168 pKa = 4.08LRR170 pKa = 11.84KK171 pKa = 9.43QIAIEE176 pKa = 3.87EE177 pKa = 4.37HH178 pKa = 5.47NLRR181 pKa = 11.84RR182 pKa = 11.84DD183 pKa = 3.36LAATARR189 pKa = 11.84QFTPANDD196 pKa = 3.48YY197 pKa = 11.22APPSKK202 pKa = 10.57AALDD206 pKa = 3.83NEE208 pKa = 4.36CWEE211 pKa = 4.83LYY213 pKa = 9.72CARR216 pKa = 11.84ARR218 pKa = 11.84DD219 pKa = 3.9AGRR222 pKa = 11.84ALPPWNAILQEE233 pKa = 4.09QATTALSNEE242 pKa = 4.7VINKK246 pKa = 8.53HH247 pKa = 5.72RR248 pKa = 11.84QDD250 pKa = 3.17FCKK253 pKa = 10.29IPSNQRR259 pKa = 11.84LLQTWAASKK268 pKa = 10.55IQDD271 pKa = 3.47LRR273 pKa = 11.84TNQEE277 pKa = 3.6FRR279 pKa = 11.84RR280 pKa = 11.84VTEE283 pKa = 4.04FEE285 pKa = 4.17RR286 pKa = 11.84FLALASGDD294 pKa = 3.28TSMASSSAPEE304 pKa = 4.04AEE306 pKa = 4.42MGQHH310 pKa = 6.98DD311 pKa = 4.1SGGEE315 pKa = 3.92GGEE318 pKa = 4.15SEE320 pKa = 4.35EE321 pKa = 4.64EE322 pKa = 4.17EE323 pKa = 4.49RR324 pKa = 11.84EE325 pKa = 4.11HH326 pKa = 6.13TAAEE330 pKa = 4.26HH331 pKa = 6.25ARR333 pKa = 11.84GGPSEE338 pKa = 4.21QPDD341 pKa = 3.57AEE343 pKa = 4.54GDD345 pKa = 3.8QEE347 pKa = 4.02QTPARR352 pKa = 11.84KK353 pKa = 9.3KK354 pKa = 10.23RR355 pKa = 11.84RR356 pKa = 11.84SKK358 pKa = 10.65RR359 pKa = 11.84LRR361 pKa = 11.84NFPSRR366 pKa = 11.84HH367 pKa = 4.39QRR369 pKa = 11.84SHH371 pKa = 6.25SGSKK375 pKa = 9.16KK376 pKa = 9.54HH377 pKa = 6.09RR378 pKa = 11.84KK379 pKa = 8.65KK380 pKa = 10.46PNRR383 pKa = 11.84RR384 pKa = 11.84DD385 pKa = 3.26SDD387 pKa = 3.71GQEE390 pKa = 3.6
MM1 pKa = 7.62ASSSGPDD8 pKa = 3.11PTQYY12 pKa = 11.62LDD14 pKa = 3.09ITLLSDD20 pKa = 3.88PVAEE24 pKa = 4.38SVSLQDD30 pKa = 4.33AVQHH34 pKa = 6.0LASIGVRR41 pKa = 11.84VEE43 pKa = 3.59RR44 pKa = 11.84FTRR47 pKa = 11.84DD48 pKa = 2.87SIHH51 pKa = 6.77AMRR54 pKa = 11.84MSVPAYY60 pKa = 9.43VKK62 pKa = 10.04EE63 pKa = 3.64IRR65 pKa = 11.84ILGNISDD72 pKa = 4.59RR73 pKa = 11.84EE74 pKa = 4.01LLKK77 pKa = 11.05NVMLQGVKK85 pKa = 10.78NNVLSLPEE93 pKa = 4.53DD94 pKa = 3.56VTPQMVLSFARR105 pKa = 11.84WLKK108 pKa = 9.84RR109 pKa = 11.84GAGARR114 pKa = 11.84VLADD118 pKa = 3.35QQHH121 pKa = 6.27LLKK124 pKa = 10.42INKK127 pKa = 9.06KK128 pKa = 10.14AVGDD132 pKa = 3.92STPDD136 pKa = 2.91IVAFAHH142 pKa = 7.2LLDD145 pKa = 4.1QQVQDD150 pKa = 3.73LTNAKK155 pKa = 9.96KK156 pKa = 10.16RR157 pKa = 11.84VQHH160 pKa = 6.03SSQVRR165 pKa = 11.84IDD167 pKa = 3.75EE168 pKa = 4.08LRR170 pKa = 11.84KK171 pKa = 9.43QIAIEE176 pKa = 3.87EE177 pKa = 4.37HH178 pKa = 5.47NLRR181 pKa = 11.84RR182 pKa = 11.84DD183 pKa = 3.36LAATARR189 pKa = 11.84QFTPANDD196 pKa = 3.48YY197 pKa = 11.22APPSKK202 pKa = 10.57AALDD206 pKa = 3.83NEE208 pKa = 4.36CWEE211 pKa = 4.83LYY213 pKa = 9.72CARR216 pKa = 11.84ARR218 pKa = 11.84DD219 pKa = 3.9AGRR222 pKa = 11.84ALPPWNAILQEE233 pKa = 4.09QATTALSNEE242 pKa = 4.7VINKK246 pKa = 8.53HH247 pKa = 5.72RR248 pKa = 11.84QDD250 pKa = 3.17FCKK253 pKa = 10.29IPSNQRR259 pKa = 11.84LLQTWAASKK268 pKa = 10.55IQDD271 pKa = 3.47LRR273 pKa = 11.84TNQEE277 pKa = 3.6FRR279 pKa = 11.84RR280 pKa = 11.84VTEE283 pKa = 4.04FEE285 pKa = 4.17RR286 pKa = 11.84FLALASGDD294 pKa = 3.28TSMASSSAPEE304 pKa = 4.04AEE306 pKa = 4.42MGQHH310 pKa = 6.98DD311 pKa = 4.1SGGEE315 pKa = 3.92GGEE318 pKa = 4.15SEE320 pKa = 4.35EE321 pKa = 4.64EE322 pKa = 4.17EE323 pKa = 4.49RR324 pKa = 11.84EE325 pKa = 4.11HH326 pKa = 6.13TAAEE330 pKa = 4.26HH331 pKa = 6.25ARR333 pKa = 11.84GGPSEE338 pKa = 4.21QPDD341 pKa = 3.57AEE343 pKa = 4.54GDD345 pKa = 3.8QEE347 pKa = 4.02QTPARR352 pKa = 11.84KK353 pKa = 9.3KK354 pKa = 10.23RR355 pKa = 11.84RR356 pKa = 11.84SKK358 pKa = 10.65RR359 pKa = 11.84LRR361 pKa = 11.84NFPSRR366 pKa = 11.84HH367 pKa = 4.39QRR369 pKa = 11.84SHH371 pKa = 6.25SGSKK375 pKa = 9.16KK376 pKa = 9.54HH377 pKa = 6.09RR378 pKa = 11.84KK379 pKa = 8.65KK380 pKa = 10.46PNRR383 pKa = 11.84RR384 pKa = 11.84DD385 pKa = 3.26SDD387 pKa = 3.71GQEE390 pKa = 3.6
Molecular weight: 43.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5MGZ5|A0A2H5MGZ5_9VIRU ORF1+2p OS=Allium cepa amalgavirus 2 OX=2058779 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.62ASSSGPDD8 pKa = 3.11PTQYY12 pKa = 11.62LDD14 pKa = 3.09ITLLSDD20 pKa = 3.88PVAEE24 pKa = 4.38SVSLQDD30 pKa = 4.33AVQHH34 pKa = 6.0LASIGVRR41 pKa = 11.84VEE43 pKa = 3.59RR44 pKa = 11.84FTRR47 pKa = 11.84DD48 pKa = 2.87SIHH51 pKa = 6.77AMRR54 pKa = 11.84MSVPAYY60 pKa = 9.43VKK62 pKa = 10.04EE63 pKa = 3.64IRR65 pKa = 11.84ILGNISDD72 pKa = 4.59RR73 pKa = 11.84EE74 pKa = 4.01LLKK77 pKa = 11.05NVMLQGVKK85 pKa = 10.78NNVLSLPEE93 pKa = 4.53DD94 pKa = 3.56VTPQMVLSFARR105 pKa = 11.84WLKK108 pKa = 9.84RR109 pKa = 11.84GAGARR114 pKa = 11.84VLADD118 pKa = 3.35QQHH121 pKa = 6.27LLKK124 pKa = 10.42INKK127 pKa = 9.06KK128 pKa = 10.14AVGDD132 pKa = 3.92STPDD136 pKa = 2.91IVAFAHH142 pKa = 7.2LLDD145 pKa = 4.1QQVQDD150 pKa = 3.73LTNAKK155 pKa = 9.96KK156 pKa = 10.16RR157 pKa = 11.84VQHH160 pKa = 6.03SSQVRR165 pKa = 11.84IDD167 pKa = 3.75EE168 pKa = 4.08LRR170 pKa = 11.84KK171 pKa = 9.43QIAIEE176 pKa = 3.87EE177 pKa = 4.37HH178 pKa = 5.47NLRR181 pKa = 11.84RR182 pKa = 11.84DD183 pKa = 3.36LAATARR189 pKa = 11.84QFTPANDD196 pKa = 3.48YY197 pKa = 11.22APPSKK202 pKa = 10.57AALDD206 pKa = 3.83NEE208 pKa = 4.36CWEE211 pKa = 4.83LYY213 pKa = 9.72CARR216 pKa = 11.84ARR218 pKa = 11.84DD219 pKa = 3.9AGRR222 pKa = 11.84ALPPWNAILQEE233 pKa = 4.09QATTALSNEE242 pKa = 4.7VINKK246 pKa = 8.53HH247 pKa = 5.72RR248 pKa = 11.84QDD250 pKa = 3.17FCKK253 pKa = 10.29IPSNQRR259 pKa = 11.84LLQTWAASKK268 pKa = 10.55IQDD271 pKa = 3.47LRR273 pKa = 11.84TNQEE277 pKa = 3.95FVASQSSNDD286 pKa = 3.03SWLSQVEE293 pKa = 4.12THH295 pKa = 6.42LWRR298 pKa = 11.84LPLPLRR304 pKa = 11.84LKK306 pKa = 8.91WANTIPVGKK315 pKa = 9.98VVSRR319 pKa = 11.84KK320 pKa = 9.26RR321 pKa = 11.84RR322 pKa = 11.84NGNILLRR329 pKa = 11.84SMLGADD335 pKa = 4.28LLNNPMQRR343 pKa = 11.84GIKK346 pKa = 9.71NKK348 pKa = 9.96PPLARR353 pKa = 11.84NAEE356 pKa = 4.12VSASVTSLLDD366 pKa = 3.04INARR370 pKa = 11.84ILVLRR375 pKa = 11.84SIEE378 pKa = 3.93RR379 pKa = 11.84SPTVGIPTARR389 pKa = 11.84SRR391 pKa = 11.84FEE393 pKa = 3.61GAVRR397 pKa = 11.84KK398 pKa = 10.18VIGGGEE404 pKa = 4.12MIDD407 pKa = 3.02WRR409 pKa = 11.84TVSNQYY415 pKa = 10.46RR416 pKa = 11.84GGGCFSDD423 pKa = 5.76AILLLADD430 pKa = 4.07ARR432 pKa = 11.84TDD434 pKa = 3.49EE435 pKa = 4.78PGKK438 pKa = 10.29FLPDD442 pKa = 3.85YY443 pKa = 7.85FTLWKK448 pKa = 10.7ARR450 pKa = 11.84DD451 pKa = 3.7ILRR454 pKa = 11.84LPSDD458 pKa = 4.01LKK460 pKa = 11.23VPCNRR465 pKa = 11.84QALKK469 pKa = 10.9VSNFNNDD476 pKa = 2.37ATAGPFFRR484 pKa = 11.84AYY486 pKa = 10.22GIKK489 pKa = 9.94SKK491 pKa = 11.34YY492 pKa = 9.28GMRR495 pKa = 11.84GLLEE499 pKa = 4.27DD500 pKa = 4.93FAWEE504 pKa = 4.76CYY506 pKa = 10.41SSFVDD511 pKa = 3.49NGGDD515 pKa = 3.48VSFLPFVASRR525 pKa = 11.84VGFRR529 pKa = 11.84TKK531 pKa = 10.5LVSQEE536 pKa = 3.55EE537 pKa = 4.13AFIRR541 pKa = 11.84FSKK544 pKa = 10.62NKK546 pKa = 10.31AIGRR550 pKa = 11.84CVMMLDD556 pKa = 4.91AIEE559 pKa = 4.28QMFSSPLYY567 pKa = 10.21NVLSKK572 pKa = 10.01LTADD576 pKa = 4.45LRR578 pKa = 11.84FDD580 pKa = 3.75PASGFRR586 pKa = 11.84NTIVRR591 pKa = 11.84ASSDD595 pKa = 3.05WAKK598 pKa = 9.69MWEE601 pKa = 4.3EE602 pKa = 3.87VKK604 pKa = 10.39KK605 pKa = 10.72AQVIVEE611 pKa = 4.28LDD613 pKa = 2.9WKK615 pKa = 10.85KK616 pKa = 10.69FDD618 pKa = 4.23RR619 pKa = 11.84EE620 pKa = 4.13RR621 pKa = 11.84PTEE624 pKa = 4.25DD625 pKa = 3.38LAFMIDD631 pKa = 3.86VIISCFKK638 pKa = 10.43PEE640 pKa = 3.94NDD642 pKa = 3.7RR643 pKa = 11.84EE644 pKa = 4.18RR645 pKa = 11.84LFLLAYY651 pKa = 7.69KK652 pKa = 10.61TMMQRR657 pKa = 11.84CLIEE661 pKa = 4.33RR662 pKa = 11.84FFVTDD667 pKa = 3.34CGGVFKK673 pKa = 11.33VEE675 pKa = 4.04GMVPSGSLWTGWVDD689 pKa = 3.2TALNILYY696 pKa = 10.45LNAALLSLGFNMTEE710 pKa = 4.18VFPKK714 pKa = 10.71CAGDD718 pKa = 4.12DD719 pKa = 3.7NLTLFMRR726 pKa = 11.84DD727 pKa = 3.04VEE729 pKa = 4.26NRR731 pKa = 11.84RR732 pKa = 11.84LLALKK737 pKa = 10.44DD738 pKa = 3.81RR739 pKa = 11.84LNSWFRR745 pKa = 11.84AGIEE749 pKa = 4.07AEE751 pKa = 4.38DD752 pKa = 4.26FLIHH756 pKa = 6.84RR757 pKa = 11.84PPFFVTRR764 pKa = 11.84EE765 pKa = 3.72QAVFPRR771 pKa = 11.84GTDD774 pKa = 3.45LTKK777 pKa = 10.18GTSKK781 pKa = 10.73IIGNAVWVPIEE792 pKa = 4.08GEE794 pKa = 4.08MVIDD798 pKa = 3.64QEE800 pKa = 4.62AGRR803 pKa = 11.84SHH805 pKa = 6.87RR806 pKa = 11.84WQYY809 pKa = 9.78VFRR812 pKa = 11.84GRR814 pKa = 11.84PKK816 pKa = 10.24FLSAYY821 pKa = 7.8WLEE824 pKa = 4.52DD825 pKa = 3.06GRR827 pKa = 11.84PIRR830 pKa = 11.84PTHH833 pKa = 6.13INSEE837 pKa = 4.08KK838 pKa = 10.75LLFPEE843 pKa = 6.17GIHH846 pKa = 7.39DD847 pKa = 5.6SIDD850 pKa = 3.47TYY852 pKa = 11.03EE853 pKa = 4.18AAVLSMVVDD862 pKa = 3.74NPFNHH867 pKa = 6.78HH868 pKa = 6.66NINHH872 pKa = 5.59CMHH875 pKa = 6.94RR876 pKa = 11.84FVICEE881 pKa = 3.66QVKK884 pKa = 9.08RR885 pKa = 11.84QARR888 pKa = 11.84MGLDD892 pKa = 3.97PIDD895 pKa = 4.17ILWLSRR901 pKa = 11.84FRR903 pKa = 11.84SSVGEE908 pKa = 3.91EE909 pKa = 3.93VPYY912 pKa = 10.93PMIASWRR919 pKa = 11.84RR920 pKa = 11.84QDD922 pKa = 2.73TWVDD926 pKa = 3.43MEE928 pKa = 4.13QLPFVRR934 pKa = 11.84DD935 pKa = 3.42YY936 pKa = 11.76VRR938 pKa = 11.84DD939 pKa = 3.58FRR941 pKa = 11.84EE942 pKa = 4.13FVSGVTSLYY951 pKa = 10.63SRR953 pKa = 11.84QSTGGLDD960 pKa = 3.05SWKK963 pKa = 8.51FTSIIRR969 pKa = 11.84GEE971 pKa = 4.35TEE973 pKa = 3.53FGEE976 pKa = 4.67GQFGNDD982 pKa = 2.67IDD984 pKa = 5.63DD985 pKa = 4.76WISWMYY991 pKa = 9.88RR992 pKa = 11.84NPMTKK997 pKa = 8.74YY998 pKa = 9.55LRR1000 pKa = 11.84PIRR1003 pKa = 11.84RR1004 pKa = 11.84FRR1006 pKa = 11.84AVGEE1010 pKa = 3.94EE1011 pKa = 3.86VRR1013 pKa = 11.84PDD1015 pKa = 3.51RR1016 pKa = 11.84EE1017 pKa = 4.17LSGSLQRR1024 pKa = 11.84CIGIYY1029 pKa = 9.41RR1030 pKa = 11.84QIRR1033 pKa = 11.84GTQEE1037 pKa = 3.5NSSSEE1042 pKa = 3.78NFAIFLSNVLRR1053 pKa = 11.84QTRR1056 pKa = 11.84SYY1058 pKa = 11.09RR1059 pKa = 11.84DD1060 pKa = 3.55ANAHH1064 pKa = 5.57PP1065 pKa = 4.36
MM1 pKa = 7.62ASSSGPDD8 pKa = 3.11PTQYY12 pKa = 11.62LDD14 pKa = 3.09ITLLSDD20 pKa = 3.88PVAEE24 pKa = 4.38SVSLQDD30 pKa = 4.33AVQHH34 pKa = 6.0LASIGVRR41 pKa = 11.84VEE43 pKa = 3.59RR44 pKa = 11.84FTRR47 pKa = 11.84DD48 pKa = 2.87SIHH51 pKa = 6.77AMRR54 pKa = 11.84MSVPAYY60 pKa = 9.43VKK62 pKa = 10.04EE63 pKa = 3.64IRR65 pKa = 11.84ILGNISDD72 pKa = 4.59RR73 pKa = 11.84EE74 pKa = 4.01LLKK77 pKa = 11.05NVMLQGVKK85 pKa = 10.78NNVLSLPEE93 pKa = 4.53DD94 pKa = 3.56VTPQMVLSFARR105 pKa = 11.84WLKK108 pKa = 9.84RR109 pKa = 11.84GAGARR114 pKa = 11.84VLADD118 pKa = 3.35QQHH121 pKa = 6.27LLKK124 pKa = 10.42INKK127 pKa = 9.06KK128 pKa = 10.14AVGDD132 pKa = 3.92STPDD136 pKa = 2.91IVAFAHH142 pKa = 7.2LLDD145 pKa = 4.1QQVQDD150 pKa = 3.73LTNAKK155 pKa = 9.96KK156 pKa = 10.16RR157 pKa = 11.84VQHH160 pKa = 6.03SSQVRR165 pKa = 11.84IDD167 pKa = 3.75EE168 pKa = 4.08LRR170 pKa = 11.84KK171 pKa = 9.43QIAIEE176 pKa = 3.87EE177 pKa = 4.37HH178 pKa = 5.47NLRR181 pKa = 11.84RR182 pKa = 11.84DD183 pKa = 3.36LAATARR189 pKa = 11.84QFTPANDD196 pKa = 3.48YY197 pKa = 11.22APPSKK202 pKa = 10.57AALDD206 pKa = 3.83NEE208 pKa = 4.36CWEE211 pKa = 4.83LYY213 pKa = 9.72CARR216 pKa = 11.84ARR218 pKa = 11.84DD219 pKa = 3.9AGRR222 pKa = 11.84ALPPWNAILQEE233 pKa = 4.09QATTALSNEE242 pKa = 4.7VINKK246 pKa = 8.53HH247 pKa = 5.72RR248 pKa = 11.84QDD250 pKa = 3.17FCKK253 pKa = 10.29IPSNQRR259 pKa = 11.84LLQTWAASKK268 pKa = 10.55IQDD271 pKa = 3.47LRR273 pKa = 11.84TNQEE277 pKa = 3.95FVASQSSNDD286 pKa = 3.03SWLSQVEE293 pKa = 4.12THH295 pKa = 6.42LWRR298 pKa = 11.84LPLPLRR304 pKa = 11.84LKK306 pKa = 8.91WANTIPVGKK315 pKa = 9.98VVSRR319 pKa = 11.84KK320 pKa = 9.26RR321 pKa = 11.84RR322 pKa = 11.84NGNILLRR329 pKa = 11.84SMLGADD335 pKa = 4.28LLNNPMQRR343 pKa = 11.84GIKK346 pKa = 9.71NKK348 pKa = 9.96PPLARR353 pKa = 11.84NAEE356 pKa = 4.12VSASVTSLLDD366 pKa = 3.04INARR370 pKa = 11.84ILVLRR375 pKa = 11.84SIEE378 pKa = 3.93RR379 pKa = 11.84SPTVGIPTARR389 pKa = 11.84SRR391 pKa = 11.84FEE393 pKa = 3.61GAVRR397 pKa = 11.84KK398 pKa = 10.18VIGGGEE404 pKa = 4.12MIDD407 pKa = 3.02WRR409 pKa = 11.84TVSNQYY415 pKa = 10.46RR416 pKa = 11.84GGGCFSDD423 pKa = 5.76AILLLADD430 pKa = 4.07ARR432 pKa = 11.84TDD434 pKa = 3.49EE435 pKa = 4.78PGKK438 pKa = 10.29FLPDD442 pKa = 3.85YY443 pKa = 7.85FTLWKK448 pKa = 10.7ARR450 pKa = 11.84DD451 pKa = 3.7ILRR454 pKa = 11.84LPSDD458 pKa = 4.01LKK460 pKa = 11.23VPCNRR465 pKa = 11.84QALKK469 pKa = 10.9VSNFNNDD476 pKa = 2.37ATAGPFFRR484 pKa = 11.84AYY486 pKa = 10.22GIKK489 pKa = 9.94SKK491 pKa = 11.34YY492 pKa = 9.28GMRR495 pKa = 11.84GLLEE499 pKa = 4.27DD500 pKa = 4.93FAWEE504 pKa = 4.76CYY506 pKa = 10.41SSFVDD511 pKa = 3.49NGGDD515 pKa = 3.48VSFLPFVASRR525 pKa = 11.84VGFRR529 pKa = 11.84TKK531 pKa = 10.5LVSQEE536 pKa = 3.55EE537 pKa = 4.13AFIRR541 pKa = 11.84FSKK544 pKa = 10.62NKK546 pKa = 10.31AIGRR550 pKa = 11.84CVMMLDD556 pKa = 4.91AIEE559 pKa = 4.28QMFSSPLYY567 pKa = 10.21NVLSKK572 pKa = 10.01LTADD576 pKa = 4.45LRR578 pKa = 11.84FDD580 pKa = 3.75PASGFRR586 pKa = 11.84NTIVRR591 pKa = 11.84ASSDD595 pKa = 3.05WAKK598 pKa = 9.69MWEE601 pKa = 4.3EE602 pKa = 3.87VKK604 pKa = 10.39KK605 pKa = 10.72AQVIVEE611 pKa = 4.28LDD613 pKa = 2.9WKK615 pKa = 10.85KK616 pKa = 10.69FDD618 pKa = 4.23RR619 pKa = 11.84EE620 pKa = 4.13RR621 pKa = 11.84PTEE624 pKa = 4.25DD625 pKa = 3.38LAFMIDD631 pKa = 3.86VIISCFKK638 pKa = 10.43PEE640 pKa = 3.94NDD642 pKa = 3.7RR643 pKa = 11.84EE644 pKa = 4.18RR645 pKa = 11.84LFLLAYY651 pKa = 7.69KK652 pKa = 10.61TMMQRR657 pKa = 11.84CLIEE661 pKa = 4.33RR662 pKa = 11.84FFVTDD667 pKa = 3.34CGGVFKK673 pKa = 11.33VEE675 pKa = 4.04GMVPSGSLWTGWVDD689 pKa = 3.2TALNILYY696 pKa = 10.45LNAALLSLGFNMTEE710 pKa = 4.18VFPKK714 pKa = 10.71CAGDD718 pKa = 4.12DD719 pKa = 3.7NLTLFMRR726 pKa = 11.84DD727 pKa = 3.04VEE729 pKa = 4.26NRR731 pKa = 11.84RR732 pKa = 11.84LLALKK737 pKa = 10.44DD738 pKa = 3.81RR739 pKa = 11.84LNSWFRR745 pKa = 11.84AGIEE749 pKa = 4.07AEE751 pKa = 4.38DD752 pKa = 4.26FLIHH756 pKa = 6.84RR757 pKa = 11.84PPFFVTRR764 pKa = 11.84EE765 pKa = 3.72QAVFPRR771 pKa = 11.84GTDD774 pKa = 3.45LTKK777 pKa = 10.18GTSKK781 pKa = 10.73IIGNAVWVPIEE792 pKa = 4.08GEE794 pKa = 4.08MVIDD798 pKa = 3.64QEE800 pKa = 4.62AGRR803 pKa = 11.84SHH805 pKa = 6.87RR806 pKa = 11.84WQYY809 pKa = 9.78VFRR812 pKa = 11.84GRR814 pKa = 11.84PKK816 pKa = 10.24FLSAYY821 pKa = 7.8WLEE824 pKa = 4.52DD825 pKa = 3.06GRR827 pKa = 11.84PIRR830 pKa = 11.84PTHH833 pKa = 6.13INSEE837 pKa = 4.08KK838 pKa = 10.75LLFPEE843 pKa = 6.17GIHH846 pKa = 7.39DD847 pKa = 5.6SIDD850 pKa = 3.47TYY852 pKa = 11.03EE853 pKa = 4.18AAVLSMVVDD862 pKa = 3.74NPFNHH867 pKa = 6.78HH868 pKa = 6.66NINHH872 pKa = 5.59CMHH875 pKa = 6.94RR876 pKa = 11.84FVICEE881 pKa = 3.66QVKK884 pKa = 9.08RR885 pKa = 11.84QARR888 pKa = 11.84MGLDD892 pKa = 3.97PIDD895 pKa = 4.17ILWLSRR901 pKa = 11.84FRR903 pKa = 11.84SSVGEE908 pKa = 3.91EE909 pKa = 3.93VPYY912 pKa = 10.93PMIASWRR919 pKa = 11.84RR920 pKa = 11.84QDD922 pKa = 2.73TWVDD926 pKa = 3.43MEE928 pKa = 4.13QLPFVRR934 pKa = 11.84DD935 pKa = 3.42YY936 pKa = 11.76VRR938 pKa = 11.84DD939 pKa = 3.58FRR941 pKa = 11.84EE942 pKa = 4.13FVSGVTSLYY951 pKa = 10.63SRR953 pKa = 11.84QSTGGLDD960 pKa = 3.05SWKK963 pKa = 8.51FTSIIRR969 pKa = 11.84GEE971 pKa = 4.35TEE973 pKa = 3.53FGEE976 pKa = 4.67GQFGNDD982 pKa = 2.67IDD984 pKa = 5.63DD985 pKa = 4.76WISWMYY991 pKa = 9.88RR992 pKa = 11.84NPMTKK997 pKa = 8.74YY998 pKa = 9.55LRR1000 pKa = 11.84PIRR1003 pKa = 11.84RR1004 pKa = 11.84FRR1006 pKa = 11.84AVGEE1010 pKa = 3.94EE1011 pKa = 3.86VRR1013 pKa = 11.84PDD1015 pKa = 3.51RR1016 pKa = 11.84EE1017 pKa = 4.17LSGSLQRR1024 pKa = 11.84CIGIYY1029 pKa = 9.41RR1030 pKa = 11.84QIRR1033 pKa = 11.84GTQEE1037 pKa = 3.5NSSSEE1042 pKa = 3.78NFAIFLSNVLRR1053 pKa = 11.84QTRR1056 pKa = 11.84SYY1058 pKa = 11.09RR1059 pKa = 11.84DD1060 pKa = 3.55ANAHH1064 pKa = 5.57PP1065 pKa = 4.36
Molecular weight: 121.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1455 |
390 |
1065 |
727.5 |
82.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.247 ± 1.152 | 1.168 ± 0.203 |
6.323 ± 0.044 | 5.979 ± 0.741 |
4.261 ± 0.993 | 5.361 ± 0.249 |
2.062 ± 0.647 | 5.223 ± 0.7 |
4.674 ± 0.362 | 9.21 ± 0.38 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.337 ± 0.276 | 4.605 ± 0.255 |
4.742 ± 0.066 | 4.811 ± 1.074 |
9.003 ± 0.116 | 7.629 ± 0.423 |
4.192 ± 0.046 | 6.392 ± 0.643 |
1.993 ± 0.492 | 1.787 ± 0.387 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
