
Ancylobacter pratisalsi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Xanthobacteraceae; Ancylobacter
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
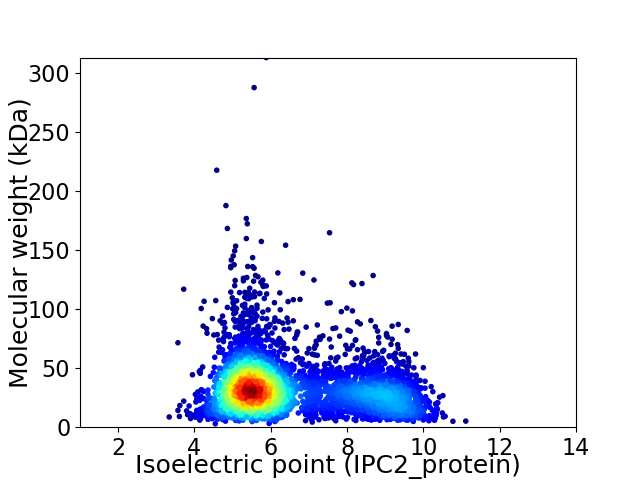
Virtual 2D-PAGE plot for 4319 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P1YK62|A0A6P1YK62_9RHIZ FAD-dependent oxidoreductase OS=Ancylobacter pratisalsi OX=1745854 GN=G3A50_07140 PE=4 SV=1
MM1 pKa = 6.79TTTTEE6 pKa = 4.09LKK8 pKa = 9.17ITLTDD13 pKa = 3.68EE14 pKa = 4.99LQDD17 pKa = 3.59TLNAGGAAVYY27 pKa = 10.08AIYY30 pKa = 10.29FNPTSGAPVIQTLFTGATSAGPATAPITVDD60 pKa = 3.22VPLTLDD66 pKa = 3.48VVSGKK71 pKa = 10.7VYY73 pKa = 10.4FLVQSPVDD81 pKa = 3.61GTLADD86 pKa = 3.55PTTFVGTQSDD96 pKa = 4.82LNWQSAEE103 pKa = 4.02THH105 pKa = 5.98NYY107 pKa = 9.98RR108 pKa = 11.84YY109 pKa = 10.56DD110 pKa = 3.64SFEE113 pKa = 3.86LTIEE117 pKa = 4.01NNINDD122 pKa = 4.44AGNLSSVEE130 pKa = 4.24GYY132 pKa = 9.99GLPMSVGVSYY142 pKa = 11.64GDD144 pKa = 3.73GSSASVGYY152 pKa = 9.87NVSGNEE158 pKa = 3.93LFHH161 pKa = 7.01ALSTMGQAQTVFPYY175 pKa = 9.58ATTAGEE181 pKa = 4.15PGLTGDD187 pKa = 4.44RR188 pKa = 11.84AGLSPSQSVGIKK200 pKa = 9.85SAAFTAGDD208 pKa = 3.26WDD210 pKa = 4.29SYY212 pKa = 11.56VDD214 pKa = 3.86YY215 pKa = 10.66LKK217 pKa = 10.96KK218 pKa = 9.91IDD220 pKa = 4.2PQTHH224 pKa = 5.77EE225 pKa = 4.48PNANAIQFAGFFDD238 pKa = 4.25GAKK241 pKa = 9.97DD242 pKa = 3.66ANGVYY247 pKa = 10.55HH248 pKa = 6.82NGGFYY253 pKa = 10.31AYY255 pKa = 10.48VLEE258 pKa = 4.09WDD260 pKa = 3.9EE261 pKa = 4.22NNGIFWLSPTDD272 pKa = 3.68DD273 pKa = 3.84SQVRR277 pKa = 11.84GYY279 pKa = 10.32IAITPEE285 pKa = 3.69QLAGNIYY292 pKa = 8.74ATEE295 pKa = 4.62GYY297 pKa = 10.34VEE299 pKa = 4.25IYY301 pKa = 9.88EE302 pKa = 4.44SKK304 pKa = 10.74SDD306 pKa = 3.88YY307 pKa = 11.37ASGDD311 pKa = 3.19AYY313 pKa = 10.58HH314 pKa = 7.39IYY316 pKa = 10.67LNTYY320 pKa = 8.31TYY322 pKa = 10.23IDD324 pKa = 3.58SSGKK328 pKa = 10.4SVTNDD333 pKa = 3.09DD334 pKa = 4.83FMDD337 pKa = 4.3AAANNQWGDD346 pKa = 3.16ILKK349 pKa = 10.63DD350 pKa = 3.41LFTGFTAGFYY360 pKa = 11.27GMTGVDD366 pKa = 3.4AQGGQVDD373 pKa = 6.05LDD375 pKa = 4.27QNWNWDD381 pKa = 3.36PTYY384 pKa = 11.46SFGANLATGEE394 pKa = 4.1APIYY398 pKa = 8.89YY399 pKa = 9.51DD400 pKa = 3.7PYY402 pKa = 10.24SAYY405 pKa = 10.39FFANSNSYY413 pKa = 10.64GSGYY417 pKa = 10.3SDD419 pKa = 3.21QLMSQYY425 pKa = 11.14SEE427 pKa = 4.18GGPLISLYY435 pKa = 10.76DD436 pKa = 3.77PSLGGSVTSIAITIFDD452 pKa = 4.95DD453 pKa = 5.04DD454 pKa = 4.33EE455 pKa = 4.73TPTGYY460 pKa = 8.79TKK462 pKa = 10.45PVIYY466 pKa = 10.52NYY468 pKa = 9.68IAPGADD474 pKa = 3.31GYY476 pKa = 9.4TPPEE480 pKa = 3.85YY481 pKa = 10.33DD482 pKa = 3.62ANSAANIVLNFANSAMILDD501 pKa = 3.93EE502 pKa = 4.18TVARR506 pKa = 11.84ITFSFQTGDD515 pKa = 3.72ASHH518 pKa = 7.28PWASVTIDD526 pKa = 3.17GSMGSLWQNWDD537 pKa = 2.8IVQEE541 pKa = 4.11KK542 pKa = 10.02DD543 pKa = 3.0GSYY546 pKa = 10.72SAVPQNPAGMQPAGTILIGKK566 pKa = 9.04LPVADD571 pKa = 5.25DD572 pKa = 4.23GVSHH576 pKa = 6.14YY577 pKa = 10.43KK578 pKa = 10.12IEE580 pKa = 4.02VGANDD585 pKa = 3.42GHH587 pKa = 6.83ASKK590 pKa = 9.64TFNLYY595 pKa = 7.14TTTNADD601 pKa = 3.93GLFLDD606 pKa = 4.72PAYY609 pKa = 10.32AGQAGAIAIDD619 pKa = 3.72GLATVSAAPATQYY632 pKa = 11.5VNTFTIDD639 pKa = 3.74FLPSTTTTIDD649 pKa = 3.24PSLLTRR655 pKa = 11.84VQSTLVPSSVVVGQVTGTDD674 pKa = 3.99PSPSPHH680 pKa = 6.37GGGFTALAGQDD691 pKa = 3.45ALNGNVVSSQHH702 pKa = 5.51AQLGFGWTGLNNATAASASWISGYY726 pKa = 7.35TNKK729 pKa = 9.4VDD731 pKa = 3.88GQSVALVTIAGAGLAPVALLADD753 pKa = 4.02IDD755 pKa = 4.35GQWVSQGKK763 pKa = 9.11SEE765 pKa = 4.21IATLGEE771 pKa = 3.87GTYY774 pKa = 9.93TVTMQQYY781 pKa = 9.09AASDD785 pKa = 3.6TAHH788 pKa = 6.98AHH790 pKa = 6.83PLTQVSSKK798 pKa = 7.75MTLTIALNEE807 pKa = 4.06MDD809 pKa = 5.69LVLAPGGAGAQLVDD823 pKa = 4.79DD824 pKa = 4.96GSGTSGNWIRR834 pKa = 11.84LEE836 pKa = 4.0TSGAALEE843 pKa = 4.77AGSSLVAYY851 pKa = 9.44AVNANGEE858 pKa = 4.36MVSRR862 pKa = 11.84DD863 pKa = 3.4GLEE866 pKa = 4.2AGASVTLQDD875 pKa = 3.75ATLGHH880 pKa = 7.05IGAIAGDD887 pKa = 4.34DD888 pKa = 3.96GSSLMFGGQSVHH900 pKa = 6.65LGAGLEE906 pKa = 4.05LRR908 pKa = 11.84FAEE911 pKa = 4.54IDD913 pKa = 3.52ASGHH917 pKa = 6.47ADD919 pKa = 4.42LSPAMNVSATPDD931 pKa = 3.17GGIQFALDD939 pKa = 3.61GFVLQASVEE948 pKa = 4.14NSLDD952 pKa = 3.4AAAMIAGNQRR962 pKa = 11.84ASDD965 pKa = 4.03TPWVYY970 pKa = 10.94LEE972 pKa = 5.06HH973 pKa = 7.29GDD975 pKa = 4.97LIQFEE980 pKa = 4.63IAGSSANTNTLGFVRR995 pKa = 11.84IDD997 pKa = 3.31VDD999 pKa = 3.67PATGDD1004 pKa = 3.13WSVGGVAYY1012 pKa = 10.49GDD1014 pKa = 3.12TDD1016 pKa = 3.66AFHH1019 pKa = 7.26DD1020 pKa = 4.3AVRR1023 pKa = 11.84GALDD1027 pKa = 5.0DD1028 pKa = 4.21GFLYY1032 pKa = 10.14QQGGNFQVTDD1042 pKa = 3.21EE1043 pKa = 4.52WEE1045 pKa = 3.97VAGASGYY1052 pKa = 8.32YY1053 pKa = 10.47APVLLTQDD1061 pKa = 3.32GEE1063 pKa = 4.71TFVIGNANDD1072 pKa = 3.66GGNDD1076 pKa = 3.83YY1077 pKa = 11.22IRR1079 pKa = 11.84MFGEE1083 pKa = 3.85NTFGIEE1089 pKa = 4.83DD1090 pKa = 3.86LTASAGSDD1098 pKa = 3.17FDD1100 pKa = 4.64YY1101 pKa = 11.45NDD1103 pKa = 3.21MVVRR1107 pKa = 11.84LLPSEE1112 pKa = 4.35EE1113 pKa = 4.14LLSS1116 pKa = 3.86
MM1 pKa = 6.79TTTTEE6 pKa = 4.09LKK8 pKa = 9.17ITLTDD13 pKa = 3.68EE14 pKa = 4.99LQDD17 pKa = 3.59TLNAGGAAVYY27 pKa = 10.08AIYY30 pKa = 10.29FNPTSGAPVIQTLFTGATSAGPATAPITVDD60 pKa = 3.22VPLTLDD66 pKa = 3.48VVSGKK71 pKa = 10.7VYY73 pKa = 10.4FLVQSPVDD81 pKa = 3.61GTLADD86 pKa = 3.55PTTFVGTQSDD96 pKa = 4.82LNWQSAEE103 pKa = 4.02THH105 pKa = 5.98NYY107 pKa = 9.98RR108 pKa = 11.84YY109 pKa = 10.56DD110 pKa = 3.64SFEE113 pKa = 3.86LTIEE117 pKa = 4.01NNINDD122 pKa = 4.44AGNLSSVEE130 pKa = 4.24GYY132 pKa = 9.99GLPMSVGVSYY142 pKa = 11.64GDD144 pKa = 3.73GSSASVGYY152 pKa = 9.87NVSGNEE158 pKa = 3.93LFHH161 pKa = 7.01ALSTMGQAQTVFPYY175 pKa = 9.58ATTAGEE181 pKa = 4.15PGLTGDD187 pKa = 4.44RR188 pKa = 11.84AGLSPSQSVGIKK200 pKa = 9.85SAAFTAGDD208 pKa = 3.26WDD210 pKa = 4.29SYY212 pKa = 11.56VDD214 pKa = 3.86YY215 pKa = 10.66LKK217 pKa = 10.96KK218 pKa = 9.91IDD220 pKa = 4.2PQTHH224 pKa = 5.77EE225 pKa = 4.48PNANAIQFAGFFDD238 pKa = 4.25GAKK241 pKa = 9.97DD242 pKa = 3.66ANGVYY247 pKa = 10.55HH248 pKa = 6.82NGGFYY253 pKa = 10.31AYY255 pKa = 10.48VLEE258 pKa = 4.09WDD260 pKa = 3.9EE261 pKa = 4.22NNGIFWLSPTDD272 pKa = 3.68DD273 pKa = 3.84SQVRR277 pKa = 11.84GYY279 pKa = 10.32IAITPEE285 pKa = 3.69QLAGNIYY292 pKa = 8.74ATEE295 pKa = 4.62GYY297 pKa = 10.34VEE299 pKa = 4.25IYY301 pKa = 9.88EE302 pKa = 4.44SKK304 pKa = 10.74SDD306 pKa = 3.88YY307 pKa = 11.37ASGDD311 pKa = 3.19AYY313 pKa = 10.58HH314 pKa = 7.39IYY316 pKa = 10.67LNTYY320 pKa = 8.31TYY322 pKa = 10.23IDD324 pKa = 3.58SSGKK328 pKa = 10.4SVTNDD333 pKa = 3.09DD334 pKa = 4.83FMDD337 pKa = 4.3AAANNQWGDD346 pKa = 3.16ILKK349 pKa = 10.63DD350 pKa = 3.41LFTGFTAGFYY360 pKa = 11.27GMTGVDD366 pKa = 3.4AQGGQVDD373 pKa = 6.05LDD375 pKa = 4.27QNWNWDD381 pKa = 3.36PTYY384 pKa = 11.46SFGANLATGEE394 pKa = 4.1APIYY398 pKa = 8.89YY399 pKa = 9.51DD400 pKa = 3.7PYY402 pKa = 10.24SAYY405 pKa = 10.39FFANSNSYY413 pKa = 10.64GSGYY417 pKa = 10.3SDD419 pKa = 3.21QLMSQYY425 pKa = 11.14SEE427 pKa = 4.18GGPLISLYY435 pKa = 10.76DD436 pKa = 3.77PSLGGSVTSIAITIFDD452 pKa = 4.95DD453 pKa = 5.04DD454 pKa = 4.33EE455 pKa = 4.73TPTGYY460 pKa = 8.79TKK462 pKa = 10.45PVIYY466 pKa = 10.52NYY468 pKa = 9.68IAPGADD474 pKa = 3.31GYY476 pKa = 9.4TPPEE480 pKa = 3.85YY481 pKa = 10.33DD482 pKa = 3.62ANSAANIVLNFANSAMILDD501 pKa = 3.93EE502 pKa = 4.18TVARR506 pKa = 11.84ITFSFQTGDD515 pKa = 3.72ASHH518 pKa = 7.28PWASVTIDD526 pKa = 3.17GSMGSLWQNWDD537 pKa = 2.8IVQEE541 pKa = 4.11KK542 pKa = 10.02DD543 pKa = 3.0GSYY546 pKa = 10.72SAVPQNPAGMQPAGTILIGKK566 pKa = 9.04LPVADD571 pKa = 5.25DD572 pKa = 4.23GVSHH576 pKa = 6.14YY577 pKa = 10.43KK578 pKa = 10.12IEE580 pKa = 4.02VGANDD585 pKa = 3.42GHH587 pKa = 6.83ASKK590 pKa = 9.64TFNLYY595 pKa = 7.14TTTNADD601 pKa = 3.93GLFLDD606 pKa = 4.72PAYY609 pKa = 10.32AGQAGAIAIDD619 pKa = 3.72GLATVSAAPATQYY632 pKa = 11.5VNTFTIDD639 pKa = 3.74FLPSTTTTIDD649 pKa = 3.24PSLLTRR655 pKa = 11.84VQSTLVPSSVVVGQVTGTDD674 pKa = 3.99PSPSPHH680 pKa = 6.37GGGFTALAGQDD691 pKa = 3.45ALNGNVVSSQHH702 pKa = 5.51AQLGFGWTGLNNATAASASWISGYY726 pKa = 7.35TNKK729 pKa = 9.4VDD731 pKa = 3.88GQSVALVTIAGAGLAPVALLADD753 pKa = 4.02IDD755 pKa = 4.35GQWVSQGKK763 pKa = 9.11SEE765 pKa = 4.21IATLGEE771 pKa = 3.87GTYY774 pKa = 9.93TVTMQQYY781 pKa = 9.09AASDD785 pKa = 3.6TAHH788 pKa = 6.98AHH790 pKa = 6.83PLTQVSSKK798 pKa = 7.75MTLTIALNEE807 pKa = 4.06MDD809 pKa = 5.69LVLAPGGAGAQLVDD823 pKa = 4.79DD824 pKa = 4.96GSGTSGNWIRR834 pKa = 11.84LEE836 pKa = 4.0TSGAALEE843 pKa = 4.77AGSSLVAYY851 pKa = 9.44AVNANGEE858 pKa = 4.36MVSRR862 pKa = 11.84DD863 pKa = 3.4GLEE866 pKa = 4.2AGASVTLQDD875 pKa = 3.75ATLGHH880 pKa = 7.05IGAIAGDD887 pKa = 4.34DD888 pKa = 3.96GSSLMFGGQSVHH900 pKa = 6.65LGAGLEE906 pKa = 4.05LRR908 pKa = 11.84FAEE911 pKa = 4.54IDD913 pKa = 3.52ASGHH917 pKa = 6.47ADD919 pKa = 4.42LSPAMNVSATPDD931 pKa = 3.17GGIQFALDD939 pKa = 3.61GFVLQASVEE948 pKa = 4.14NSLDD952 pKa = 3.4AAAMIAGNQRR962 pKa = 11.84ASDD965 pKa = 4.03TPWVYY970 pKa = 10.94LEE972 pKa = 5.06HH973 pKa = 7.29GDD975 pKa = 4.97LIQFEE980 pKa = 4.63IAGSSANTNTLGFVRR995 pKa = 11.84IDD997 pKa = 3.31VDD999 pKa = 3.67PATGDD1004 pKa = 3.13WSVGGVAYY1012 pKa = 10.49GDD1014 pKa = 3.12TDD1016 pKa = 3.66AFHH1019 pKa = 7.26DD1020 pKa = 4.3AVRR1023 pKa = 11.84GALDD1027 pKa = 5.0DD1028 pKa = 4.21GFLYY1032 pKa = 10.14QQGGNFQVTDD1042 pKa = 3.21EE1043 pKa = 4.52WEE1045 pKa = 3.97VAGASGYY1052 pKa = 8.32YY1053 pKa = 10.47APVLLTQDD1061 pKa = 3.32GEE1063 pKa = 4.71TFVIGNANDD1072 pKa = 3.66GGNDD1076 pKa = 3.83YY1077 pKa = 11.22IRR1079 pKa = 11.84MFGEE1083 pKa = 3.85NTFGIEE1089 pKa = 4.83DD1090 pKa = 3.86LTASAGSDD1098 pKa = 3.17FDD1100 pKa = 4.64YY1101 pKa = 11.45NDD1103 pKa = 3.21MVVRR1107 pKa = 11.84LLPSEE1112 pKa = 4.35EE1113 pKa = 4.14LLSS1116 pKa = 3.86
Molecular weight: 116.84 kDa
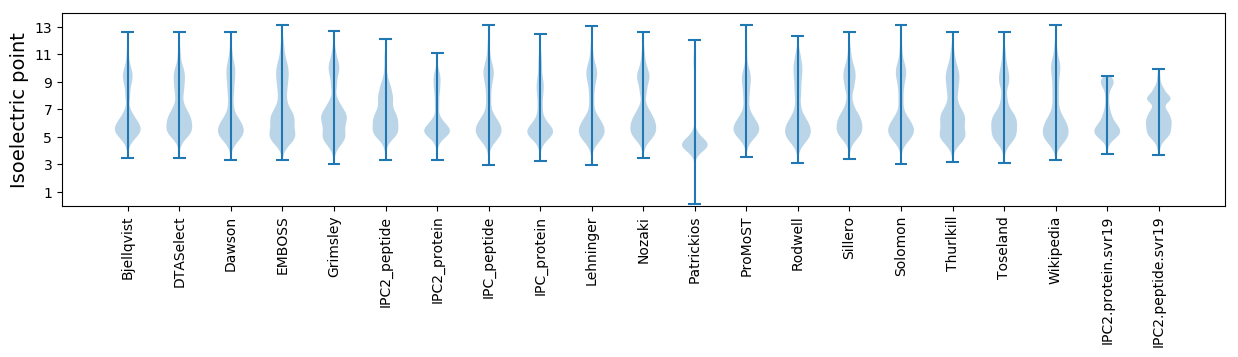
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P1YMP6|A0A6P1YMP6_9RHIZ Uncharacterized protein OS=Ancylobacter pratisalsi OX=1745854 GN=G3A50_09855 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATSNGRR28 pKa = 11.84KK29 pKa = 8.97ILNARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 4.94GRR39 pKa = 11.84KK40 pKa = 9.3RR41 pKa = 11.84LSAA44 pKa = 4.01
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.43LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.38GFRR19 pKa = 11.84ARR21 pKa = 11.84MATSNGRR28 pKa = 11.84KK29 pKa = 8.97ILNARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 4.94GRR39 pKa = 11.84KK40 pKa = 9.3RR41 pKa = 11.84LSAA44 pKa = 4.01
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1392111 |
25 |
2850 |
322.3 |
34.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.166 ± 0.057 | 0.786 ± 0.011 |
5.504 ± 0.028 | 5.632 ± 0.038 |
3.694 ± 0.026 | 8.845 ± 0.035 |
2.022 ± 0.016 | 5.014 ± 0.027 |
2.775 ± 0.032 | 10.412 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.429 ± 0.016 | 2.317 ± 0.018 |
5.522 ± 0.031 | 2.795 ± 0.021 |
7.353 ± 0.041 | 5.259 ± 0.023 |
5.268 ± 0.022 | 7.762 ± 0.027 |
1.311 ± 0.016 | 2.137 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
