
Wenzhou crab virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.26
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
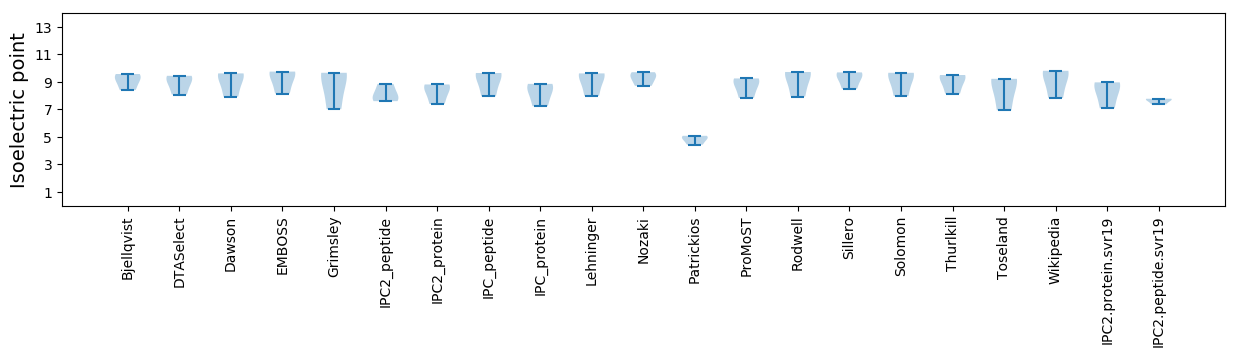
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KH85|A0A1L3KH85_9VIRU Chropara_Vmeth domain-containing protein OS=Wenzhou crab virus 4 OX=1923561 PE=4 SV=1
MM1 pKa = 7.52AGIEE5 pKa = 4.12KK6 pKa = 10.18RR7 pKa = 11.84VLAVVPKK14 pKa = 8.86PHH16 pKa = 7.59PDD18 pKa = 3.91LIRR21 pKa = 11.84GRR23 pKa = 11.84TEE25 pKa = 3.62LPRR28 pKa = 11.84TFHH31 pKa = 6.11TAASALCPTRR41 pKa = 11.84TPATADD47 pKa = 3.04TAAPYY52 pKa = 10.52LGIPPIYY59 pKa = 9.85LQPLQQTSRR68 pKa = 11.84EE69 pKa = 4.11WQEE72 pKa = 3.55AVLDD76 pKa = 3.41IARR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84KK82 pKa = 9.61VQDD85 pKa = 3.13PTLPWSVVPTLEE97 pKa = 3.95EE98 pKa = 4.66LGDD101 pKa = 4.03SSCCHH106 pKa = 7.32LGPIPDD112 pKa = 3.94RR113 pKa = 11.84RR114 pKa = 11.84DD115 pKa = 3.38KK116 pKa = 10.48LTHH119 pKa = 6.32PWFKK123 pKa = 11.29SNLHH127 pKa = 5.95RR128 pKa = 11.84LVSEE132 pKa = 4.69FWKK135 pKa = 10.78LPEE138 pKa = 4.25NQWLDD143 pKa = 3.46AADD146 pKa = 4.15PLPFEE151 pKa = 4.28EE152 pKa = 4.53WVLRR156 pKa = 11.84FPLARR161 pKa = 11.84RR162 pKa = 11.84PLLRR166 pKa = 11.84QARR169 pKa = 11.84EE170 pKa = 3.82EE171 pKa = 4.13VHH173 pKa = 5.82QHH175 pKa = 5.49GLTDD179 pKa = 3.68KK180 pKa = 10.24DD181 pKa = 2.92AVMAVFIKK189 pKa = 10.19TEE191 pKa = 3.96TSTTATDD198 pKa = 3.68PRR200 pKa = 11.84NISPRR205 pKa = 11.84QLKK208 pKa = 9.59FLSVLGPYY216 pKa = 8.94VAAIEE221 pKa = 4.27KK222 pKa = 9.03QAKK225 pKa = 7.95RR226 pKa = 11.84CSYY229 pKa = 9.9LVKK232 pKa = 10.68GLVPEE237 pKa = 4.11QRR239 pKa = 11.84GKK241 pKa = 9.36RR242 pKa = 11.84TAQSWRR248 pKa = 11.84AEE250 pKa = 4.16VIEE253 pKa = 4.26TDD255 pKa = 4.29FSRR258 pKa = 11.84FDD260 pKa = 3.38MTVSQDD266 pKa = 3.37FIEE269 pKa = 4.28IVEE272 pKa = 3.89RR273 pKa = 11.84SMFRR277 pKa = 11.84RR278 pKa = 11.84AYY280 pKa = 9.4PAGSHH285 pKa = 7.1PDD287 pKa = 3.46FDD289 pKa = 4.59SCLQMLSRR297 pKa = 11.84MTGASSLGVLYY308 pKa = 10.89SVAGTRR314 pKa = 11.84ASGDD318 pKa = 3.08AHH320 pKa = 5.82TSIANGVLNRR330 pKa = 11.84FVIWACLRR338 pKa = 11.84NLDD341 pKa = 4.1PSTWDD346 pKa = 3.46SFHH349 pKa = 7.08EE350 pKa = 4.64GDD352 pKa = 5.63DD353 pKa = 5.25GIIHH357 pKa = 7.13CDD359 pKa = 3.3KK360 pKa = 10.56PVKK363 pKa = 10.17QDD365 pKa = 2.56ICDD368 pKa = 3.89YY369 pKa = 11.32LNFAALLGFKK379 pKa = 10.39LKK381 pKa = 10.3VVRR384 pKa = 11.84PLSHH388 pKa = 6.14EE389 pKa = 3.84HH390 pKa = 6.84ANFCGRR396 pKa = 11.84HH397 pKa = 4.39TCSVCHH403 pKa = 7.07KK404 pKa = 8.2EE405 pKa = 4.31FCDD408 pKa = 4.99LPRR411 pKa = 11.84TLSKK415 pKa = 10.36FHH417 pKa = 5.68VTFKK421 pKa = 10.58QGEE424 pKa = 4.33PKK426 pKa = 10.52SLLLAKK432 pKa = 10.02ALSYY436 pKa = 9.96MATDD440 pKa = 2.98NHH442 pKa = 6.05TPIISVLCQALIEE455 pKa = 4.38HH456 pKa = 7.3LSPVVSKK463 pKa = 10.87RR464 pKa = 11.84LLARR468 pKa = 11.84RR469 pKa = 11.84IAALGWWDD477 pKa = 3.77RR478 pKa = 11.84EE479 pKa = 3.99RR480 pKa = 11.84VKK482 pKa = 10.9QGMVTTQLEE491 pKa = 4.2PTACCRR497 pKa = 11.84AAVCSVYY504 pKa = 10.65DD505 pKa = 3.82WEE507 pKa = 5.25PALQLAWEE515 pKa = 4.32RR516 pKa = 11.84QLATWKK522 pKa = 11.08YY523 pKa = 9.42GVTWLQPLSVDD534 pKa = 4.49DD535 pKa = 4.36YY536 pKa = 11.65LVDD539 pKa = 3.75SDD541 pKa = 5.65AVTIYY546 pKa = 10.5QGG548 pKa = 3.06
MM1 pKa = 7.52AGIEE5 pKa = 4.12KK6 pKa = 10.18RR7 pKa = 11.84VLAVVPKK14 pKa = 8.86PHH16 pKa = 7.59PDD18 pKa = 3.91LIRR21 pKa = 11.84GRR23 pKa = 11.84TEE25 pKa = 3.62LPRR28 pKa = 11.84TFHH31 pKa = 6.11TAASALCPTRR41 pKa = 11.84TPATADD47 pKa = 3.04TAAPYY52 pKa = 10.52LGIPPIYY59 pKa = 9.85LQPLQQTSRR68 pKa = 11.84EE69 pKa = 4.11WQEE72 pKa = 3.55AVLDD76 pKa = 3.41IARR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84KK82 pKa = 9.61VQDD85 pKa = 3.13PTLPWSVVPTLEE97 pKa = 3.95EE98 pKa = 4.66LGDD101 pKa = 4.03SSCCHH106 pKa = 7.32LGPIPDD112 pKa = 3.94RR113 pKa = 11.84RR114 pKa = 11.84DD115 pKa = 3.38KK116 pKa = 10.48LTHH119 pKa = 6.32PWFKK123 pKa = 11.29SNLHH127 pKa = 5.95RR128 pKa = 11.84LVSEE132 pKa = 4.69FWKK135 pKa = 10.78LPEE138 pKa = 4.25NQWLDD143 pKa = 3.46AADD146 pKa = 4.15PLPFEE151 pKa = 4.28EE152 pKa = 4.53WVLRR156 pKa = 11.84FPLARR161 pKa = 11.84RR162 pKa = 11.84PLLRR166 pKa = 11.84QARR169 pKa = 11.84EE170 pKa = 3.82EE171 pKa = 4.13VHH173 pKa = 5.82QHH175 pKa = 5.49GLTDD179 pKa = 3.68KK180 pKa = 10.24DD181 pKa = 2.92AVMAVFIKK189 pKa = 10.19TEE191 pKa = 3.96TSTTATDD198 pKa = 3.68PRR200 pKa = 11.84NISPRR205 pKa = 11.84QLKK208 pKa = 9.59FLSVLGPYY216 pKa = 8.94VAAIEE221 pKa = 4.27KK222 pKa = 9.03QAKK225 pKa = 7.95RR226 pKa = 11.84CSYY229 pKa = 9.9LVKK232 pKa = 10.68GLVPEE237 pKa = 4.11QRR239 pKa = 11.84GKK241 pKa = 9.36RR242 pKa = 11.84TAQSWRR248 pKa = 11.84AEE250 pKa = 4.16VIEE253 pKa = 4.26TDD255 pKa = 4.29FSRR258 pKa = 11.84FDD260 pKa = 3.38MTVSQDD266 pKa = 3.37FIEE269 pKa = 4.28IVEE272 pKa = 3.89RR273 pKa = 11.84SMFRR277 pKa = 11.84RR278 pKa = 11.84AYY280 pKa = 9.4PAGSHH285 pKa = 7.1PDD287 pKa = 3.46FDD289 pKa = 4.59SCLQMLSRR297 pKa = 11.84MTGASSLGVLYY308 pKa = 10.89SVAGTRR314 pKa = 11.84ASGDD318 pKa = 3.08AHH320 pKa = 5.82TSIANGVLNRR330 pKa = 11.84FVIWACLRR338 pKa = 11.84NLDD341 pKa = 4.1PSTWDD346 pKa = 3.46SFHH349 pKa = 7.08EE350 pKa = 4.64GDD352 pKa = 5.63DD353 pKa = 5.25GIIHH357 pKa = 7.13CDD359 pKa = 3.3KK360 pKa = 10.56PVKK363 pKa = 10.17QDD365 pKa = 2.56ICDD368 pKa = 3.89YY369 pKa = 11.32LNFAALLGFKK379 pKa = 10.39LKK381 pKa = 10.3VVRR384 pKa = 11.84PLSHH388 pKa = 6.14EE389 pKa = 3.84HH390 pKa = 6.84ANFCGRR396 pKa = 11.84HH397 pKa = 4.39TCSVCHH403 pKa = 7.07KK404 pKa = 8.2EE405 pKa = 4.31FCDD408 pKa = 4.99LPRR411 pKa = 11.84TLSKK415 pKa = 10.36FHH417 pKa = 5.68VTFKK421 pKa = 10.58QGEE424 pKa = 4.33PKK426 pKa = 10.52SLLLAKK432 pKa = 10.02ALSYY436 pKa = 9.96MATDD440 pKa = 2.98NHH442 pKa = 6.05TPIISVLCQALIEE455 pKa = 4.38HH456 pKa = 7.3LSPVVSKK463 pKa = 10.87RR464 pKa = 11.84LLARR468 pKa = 11.84RR469 pKa = 11.84IAALGWWDD477 pKa = 3.77RR478 pKa = 11.84EE479 pKa = 3.99RR480 pKa = 11.84VKK482 pKa = 10.9QGMVTTQLEE491 pKa = 4.2PTACCRR497 pKa = 11.84AAVCSVYY504 pKa = 10.65DD505 pKa = 3.82WEE507 pKa = 5.25PALQLAWEE515 pKa = 4.32RR516 pKa = 11.84QLATWKK522 pKa = 11.08YY523 pKa = 9.42GVTWLQPLSVDD534 pKa = 4.49DD535 pKa = 4.36YY536 pKa = 11.65LVDD539 pKa = 3.75SDD541 pKa = 5.65AVTIYY546 pKa = 10.5QGG548 pKa = 3.06
Molecular weight: 61.96 kDa
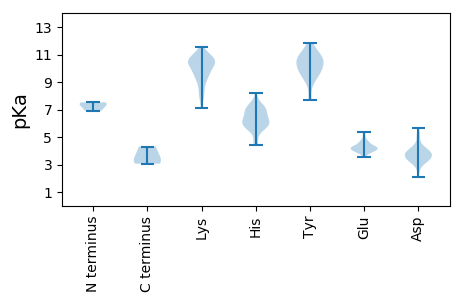
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH85|A0A1L3KH85_9VIRU Chropara_Vmeth domain-containing protein OS=Wenzhou crab virus 4 OX=1923561 PE=4 SV=1
MM1 pKa = 6.92FTLLSTLTALLVGAAVDD18 pKa = 3.51RR19 pKa = 11.84AVYY22 pKa = 10.03PDD24 pKa = 3.26VHH26 pKa = 6.57EE27 pKa = 4.59PSLYY31 pKa = 10.45EE32 pKa = 3.92RR33 pKa = 11.84TGLPLFRR40 pKa = 11.84TLQQYY45 pKa = 10.05HH46 pKa = 5.97VLDD49 pKa = 3.79VLDD52 pKa = 5.1GVGLLNILGIPDD64 pKa = 3.83DD65 pKa = 3.92FQYY68 pKa = 11.18QEE70 pKa = 4.82LAHH73 pKa = 6.2SCQTRR78 pKa = 11.84YY79 pKa = 11.22DD80 pKa = 3.66NDD82 pKa = 3.16ATTLYY87 pKa = 10.18LRR89 pKa = 11.84SWEE92 pKa = 4.27LPHH95 pKa = 7.2LSRR98 pKa = 11.84SPLPWVCQRR107 pKa = 11.84LMSRR111 pKa = 11.84GHH113 pKa = 4.92VSEE116 pKa = 4.86GRR118 pKa = 11.84VSPLILALTAPLRR131 pKa = 11.84LAHH134 pKa = 5.78WTPSIVAFSVATIGLISLCAVAALVCFIRR163 pKa = 11.84RR164 pKa = 11.84YY165 pKa = 9.87RR166 pKa = 11.84LVDD169 pKa = 3.47QVCDD173 pKa = 3.6RR174 pKa = 11.84PPFSHH179 pKa = 7.19RR180 pKa = 11.84EE181 pKa = 3.83VRR183 pKa = 11.84TLLQRR188 pKa = 11.84QIDD191 pKa = 3.94RR192 pKa = 11.84RR193 pKa = 11.84VKK195 pKa = 10.23VSDD198 pKa = 3.38VTTTPTGHH206 pKa = 6.32EE207 pKa = 3.91RR208 pKa = 11.84LAHH211 pKa = 5.62IRR213 pKa = 11.84RR214 pKa = 11.84EE215 pKa = 4.13VEE217 pKa = 4.24GACLSVLSSISSKK230 pKa = 10.69LRR232 pKa = 11.84DD233 pKa = 3.43VGGSLSRR240 pKa = 11.84NARR243 pKa = 11.84LGKK246 pKa = 9.65NLHH249 pKa = 6.1ICFPSVDD256 pKa = 3.87ALDD259 pKa = 4.34KK260 pKa = 10.91LRR262 pKa = 11.84KK263 pKa = 8.11ATNPTPDD270 pKa = 3.48NDD272 pKa = 3.9VGHH275 pKa = 6.92HH276 pKa = 6.38RR277 pKa = 11.84GEE279 pKa = 4.69DD280 pKa = 3.77CPHH283 pKa = 6.69SNRR286 pKa = 11.84PTIMTYY292 pKa = 10.76VDD294 pKa = 3.69FHH296 pKa = 8.17LPIEE300 pKa = 4.25ALTRR304 pKa = 11.84CIRR307 pKa = 11.84SPTFIVTHH315 pKa = 6.53DD316 pKa = 4.07FARR319 pKa = 11.84VLGEE323 pKa = 3.87EE324 pKa = 4.04SWFDD328 pKa = 3.73GEE330 pKa = 4.36ATVNRR335 pKa = 11.84IGDD338 pKa = 4.93FISMTTRR345 pKa = 11.84GGSSYY350 pKa = 10.33VHH352 pKa = 6.81GFHH355 pKa = 6.49SWEE358 pKa = 4.12AEE360 pKa = 3.9GVVATEE366 pKa = 4.58HH367 pKa = 6.92GVFSYY372 pKa = 10.78QRR374 pKa = 11.84VYY376 pKa = 11.13DD377 pKa = 3.66EE378 pKa = 5.38HH379 pKa = 5.77NTIVLWCVPLPGRR392 pKa = 11.84FNPGRR397 pKa = 11.84DD398 pKa = 3.41NMLTSSAGRR407 pKa = 11.84VTDD410 pKa = 4.63FDD412 pKa = 4.32LADD415 pKa = 4.06GSRR418 pKa = 11.84VTLDD422 pKa = 3.13GEE424 pKa = 4.39QYY426 pKa = 10.58RR427 pKa = 11.84IARR430 pKa = 11.84PGAPVRR436 pKa = 11.84HH437 pKa = 5.66VPAKK441 pKa = 9.64TILRR445 pKa = 11.84VAFQMCIMARR455 pKa = 11.84DD456 pKa = 4.14EE457 pKa = 4.01KK458 pKa = 10.47WLPNLHH464 pKa = 5.62GVLRR468 pKa = 11.84GRR470 pKa = 11.84FAADD474 pKa = 3.27KK475 pKa = 11.14VNEE478 pKa = 4.15EE479 pKa = 4.58CLLPAQQLCVEE490 pKa = 5.02LSDD493 pKa = 5.18LMAMCYY499 pKa = 9.57RR500 pKa = 11.84RR501 pKa = 11.84SAIGDD506 pKa = 3.68PDD508 pKa = 3.83SYY510 pKa = 11.81GPLARR515 pKa = 11.84PIVRR519 pKa = 11.84MYY521 pKa = 11.13LRR523 pKa = 11.84VLSCLPYY530 pKa = 9.82TAEE533 pKa = 4.02QFCTRR538 pKa = 11.84LVLNFLAMLHH548 pKa = 6.26KK549 pKa = 10.36PGPTLPTSWAWNTVKK564 pKa = 10.48VPTYY568 pKa = 9.52EE569 pKa = 4.14VHH571 pKa = 6.37WDD573 pKa = 3.54QVASKK578 pKa = 10.73LVDD581 pKa = 3.42AQNGLRR587 pKa = 11.84VKK589 pKa = 10.65QPFRR593 pKa = 11.84NSGAVAGAPAPNQPCAPPRR612 pKa = 11.84PDD614 pKa = 3.09QRR616 pKa = 11.84KK617 pKa = 9.45SNGRR621 pKa = 11.84DD622 pKa = 2.98RR623 pKa = 11.84KK624 pKa = 9.39EE625 pKa = 4.12GPGGGSKK632 pKa = 7.14TTSGPHH638 pKa = 4.63QGKK641 pKa = 9.82DD642 pKa = 2.62RR643 pKa = 11.84VAQNVSHH650 pKa = 6.81GSKK653 pKa = 10.56CPVSNTYY660 pKa = 9.69PRR662 pKa = 11.84NCRR665 pKa = 11.84HH666 pKa = 5.86CRR668 pKa = 11.84AVSRR672 pKa = 11.84DD673 pKa = 3.71TAHH676 pKa = 7.45LPATTPANQPRR687 pKa = 11.84VAGGSAGHH695 pKa = 6.86RR696 pKa = 11.84PQAQGSRR703 pKa = 11.84PNSTVVSRR711 pKa = 11.84PHH713 pKa = 5.5FRR715 pKa = 11.84GARR718 pKa = 11.84RR719 pKa = 11.84QLVLPSRR726 pKa = 11.84PNPGQEE732 pKa = 3.8RR733 pKa = 11.84QIDD736 pKa = 3.73TSLVQIKK743 pKa = 7.76PTSSRR748 pKa = 11.84LRR750 pKa = 11.84VLEE753 pKa = 4.16APRR756 pKa = 11.84KK757 pKa = 8.06PVAGRR762 pKa = 11.84RR763 pKa = 11.84GSSSFF768 pKa = 3.39
MM1 pKa = 6.92FTLLSTLTALLVGAAVDD18 pKa = 3.51RR19 pKa = 11.84AVYY22 pKa = 10.03PDD24 pKa = 3.26VHH26 pKa = 6.57EE27 pKa = 4.59PSLYY31 pKa = 10.45EE32 pKa = 3.92RR33 pKa = 11.84TGLPLFRR40 pKa = 11.84TLQQYY45 pKa = 10.05HH46 pKa = 5.97VLDD49 pKa = 3.79VLDD52 pKa = 5.1GVGLLNILGIPDD64 pKa = 3.83DD65 pKa = 3.92FQYY68 pKa = 11.18QEE70 pKa = 4.82LAHH73 pKa = 6.2SCQTRR78 pKa = 11.84YY79 pKa = 11.22DD80 pKa = 3.66NDD82 pKa = 3.16ATTLYY87 pKa = 10.18LRR89 pKa = 11.84SWEE92 pKa = 4.27LPHH95 pKa = 7.2LSRR98 pKa = 11.84SPLPWVCQRR107 pKa = 11.84LMSRR111 pKa = 11.84GHH113 pKa = 4.92VSEE116 pKa = 4.86GRR118 pKa = 11.84VSPLILALTAPLRR131 pKa = 11.84LAHH134 pKa = 5.78WTPSIVAFSVATIGLISLCAVAALVCFIRR163 pKa = 11.84RR164 pKa = 11.84YY165 pKa = 9.87RR166 pKa = 11.84LVDD169 pKa = 3.47QVCDD173 pKa = 3.6RR174 pKa = 11.84PPFSHH179 pKa = 7.19RR180 pKa = 11.84EE181 pKa = 3.83VRR183 pKa = 11.84TLLQRR188 pKa = 11.84QIDD191 pKa = 3.94RR192 pKa = 11.84RR193 pKa = 11.84VKK195 pKa = 10.23VSDD198 pKa = 3.38VTTTPTGHH206 pKa = 6.32EE207 pKa = 3.91RR208 pKa = 11.84LAHH211 pKa = 5.62IRR213 pKa = 11.84RR214 pKa = 11.84EE215 pKa = 4.13VEE217 pKa = 4.24GACLSVLSSISSKK230 pKa = 10.69LRR232 pKa = 11.84DD233 pKa = 3.43VGGSLSRR240 pKa = 11.84NARR243 pKa = 11.84LGKK246 pKa = 9.65NLHH249 pKa = 6.1ICFPSVDD256 pKa = 3.87ALDD259 pKa = 4.34KK260 pKa = 10.91LRR262 pKa = 11.84KK263 pKa = 8.11ATNPTPDD270 pKa = 3.48NDD272 pKa = 3.9VGHH275 pKa = 6.92HH276 pKa = 6.38RR277 pKa = 11.84GEE279 pKa = 4.69DD280 pKa = 3.77CPHH283 pKa = 6.69SNRR286 pKa = 11.84PTIMTYY292 pKa = 10.76VDD294 pKa = 3.69FHH296 pKa = 8.17LPIEE300 pKa = 4.25ALTRR304 pKa = 11.84CIRR307 pKa = 11.84SPTFIVTHH315 pKa = 6.53DD316 pKa = 4.07FARR319 pKa = 11.84VLGEE323 pKa = 3.87EE324 pKa = 4.04SWFDD328 pKa = 3.73GEE330 pKa = 4.36ATVNRR335 pKa = 11.84IGDD338 pKa = 4.93FISMTTRR345 pKa = 11.84GGSSYY350 pKa = 10.33VHH352 pKa = 6.81GFHH355 pKa = 6.49SWEE358 pKa = 4.12AEE360 pKa = 3.9GVVATEE366 pKa = 4.58HH367 pKa = 6.92GVFSYY372 pKa = 10.78QRR374 pKa = 11.84VYY376 pKa = 11.13DD377 pKa = 3.66EE378 pKa = 5.38HH379 pKa = 5.77NTIVLWCVPLPGRR392 pKa = 11.84FNPGRR397 pKa = 11.84DD398 pKa = 3.41NMLTSSAGRR407 pKa = 11.84VTDD410 pKa = 4.63FDD412 pKa = 4.32LADD415 pKa = 4.06GSRR418 pKa = 11.84VTLDD422 pKa = 3.13GEE424 pKa = 4.39QYY426 pKa = 10.58RR427 pKa = 11.84IARR430 pKa = 11.84PGAPVRR436 pKa = 11.84HH437 pKa = 5.66VPAKK441 pKa = 9.64TILRR445 pKa = 11.84VAFQMCIMARR455 pKa = 11.84DD456 pKa = 4.14EE457 pKa = 4.01KK458 pKa = 10.47WLPNLHH464 pKa = 5.62GVLRR468 pKa = 11.84GRR470 pKa = 11.84FAADD474 pKa = 3.27KK475 pKa = 11.14VNEE478 pKa = 4.15EE479 pKa = 4.58CLLPAQQLCVEE490 pKa = 5.02LSDD493 pKa = 5.18LMAMCYY499 pKa = 9.57RR500 pKa = 11.84RR501 pKa = 11.84SAIGDD506 pKa = 3.68PDD508 pKa = 3.83SYY510 pKa = 11.81GPLARR515 pKa = 11.84PIVRR519 pKa = 11.84MYY521 pKa = 11.13LRR523 pKa = 11.84VLSCLPYY530 pKa = 9.82TAEE533 pKa = 4.02QFCTRR538 pKa = 11.84LVLNFLAMLHH548 pKa = 6.26KK549 pKa = 10.36PGPTLPTSWAWNTVKK564 pKa = 10.48VPTYY568 pKa = 9.52EE569 pKa = 4.14VHH571 pKa = 6.37WDD573 pKa = 3.54QVASKK578 pKa = 10.73LVDD581 pKa = 3.42AQNGLRR587 pKa = 11.84VKK589 pKa = 10.65QPFRR593 pKa = 11.84NSGAVAGAPAPNQPCAPPRR612 pKa = 11.84PDD614 pKa = 3.09QRR616 pKa = 11.84KK617 pKa = 9.45SNGRR621 pKa = 11.84DD622 pKa = 2.98RR623 pKa = 11.84KK624 pKa = 9.39EE625 pKa = 4.12GPGGGSKK632 pKa = 7.14TTSGPHH638 pKa = 4.63QGKK641 pKa = 9.82DD642 pKa = 2.62RR643 pKa = 11.84VAQNVSHH650 pKa = 6.81GSKK653 pKa = 10.56CPVSNTYY660 pKa = 9.69PRR662 pKa = 11.84NCRR665 pKa = 11.84HH666 pKa = 5.86CRR668 pKa = 11.84AVSRR672 pKa = 11.84DD673 pKa = 3.71TAHH676 pKa = 7.45LPATTPANQPRR687 pKa = 11.84VAGGSAGHH695 pKa = 6.86RR696 pKa = 11.84PQAQGSRR703 pKa = 11.84PNSTVVSRR711 pKa = 11.84PHH713 pKa = 5.5FRR715 pKa = 11.84GARR718 pKa = 11.84RR719 pKa = 11.84QLVLPSRR726 pKa = 11.84PNPGQEE732 pKa = 3.8RR733 pKa = 11.84QIDD736 pKa = 3.73TSLVQIKK743 pKa = 7.76PTSSRR748 pKa = 11.84LRR750 pKa = 11.84VLEE753 pKa = 4.16APRR756 pKa = 11.84KK757 pKa = 8.06PVAGRR762 pKa = 11.84RR763 pKa = 11.84GSSSFF768 pKa = 3.39
Molecular weight: 85.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1675 |
359 |
768 |
558.3 |
62.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.239 ± 0.417 | 2.328 ± 0.562 |
5.612 ± 0.23 | 4.06 ± 0.42 |
3.403 ± 0.264 | 6.09 ± 0.66 |
2.985 ± 0.842 | 3.642 ± 0.222 |
3.642 ± 0.702 | 9.493 ± 0.959 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.552 ± 0.152 | 3.104 ± 0.778 |
7.284 ± 0.231 | 4.418 ± 0.672 |
8.358 ± 1.01 | 6.985 ± 0.348 |
6.627 ± 0.565 | 8.179 ± 0.472 |
1.731 ± 0.395 | 2.269 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
