
human papillomavirus 153
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 13
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
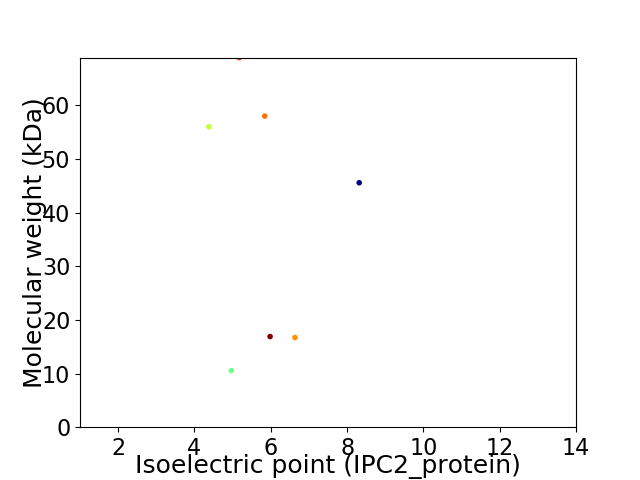
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8GMN0|G8GMN0_9PAPI Major capsid protein L1 OS=human papillomavirus 153 OX=1110710 GN=L1 PE=3 SV=1
MM1 pKa = 7.38SNNRR5 pKa = 11.84KK6 pKa = 9.02RR7 pKa = 11.84RR8 pKa = 11.84AAPIDD13 pKa = 4.13LYY15 pKa = 11.09KK16 pKa = 10.2SCQMGGDD23 pKa = 4.47CFPDD27 pKa = 3.35VQNKK31 pKa = 9.49IEE33 pKa = 4.19GTTLADD39 pKa = 3.55RR40 pKa = 11.84LLQIFSSVLYY50 pKa = 10.39FGGLGIGSGKK60 pKa = 8.44GTGGSTGYY68 pKa = 10.81KK69 pKa = 9.67PLTPGATQRR78 pKa = 11.84PSQFPQTFRR87 pKa = 11.84PTIPIDD93 pKa = 3.58PLGGADD99 pKa = 5.25VIPLDD104 pKa = 4.98IIDD107 pKa = 4.54ASTPSVVPLIDD118 pKa = 3.68GTPEE122 pKa = 3.63TTIISSGTGPTVEE135 pKa = 5.19IGDD138 pKa = 4.41LDD140 pKa = 3.68VTTTVDD146 pKa = 3.18ISATSVARR154 pKa = 11.84GDD156 pKa = 3.55HH157 pKa = 6.39PAIIDD162 pKa = 3.41ISEE165 pKa = 4.34EE166 pKa = 4.12GSSVIDD172 pKa = 3.88VQNGPPPPKK181 pKa = 10.08KK182 pKa = 10.4LQLDD186 pKa = 3.88TLLNPPEE193 pKa = 4.63TIDD196 pKa = 4.85LVTPPTTTYY205 pKa = 10.46RR206 pKa = 11.84DD207 pKa = 3.32SNINVFVDD215 pKa = 3.67THH217 pKa = 7.5ISGEE221 pKa = 4.34VIGAEE226 pKa = 4.29SIPLDD231 pKa = 3.68EE232 pKa = 5.69LSTIEE237 pKa = 3.84QFEE240 pKa = 4.29IEE242 pKa = 4.58EE243 pKa = 4.34PRR245 pKa = 11.84QTSTPEE251 pKa = 3.38ARR253 pKa = 11.84LNKK256 pKa = 10.07AIYY259 pKa = 8.93KK260 pKa = 10.57AKK262 pKa = 9.07TLYY265 pKa = 10.58NRR267 pKa = 11.84FVKK270 pKa = 10.0QVPTQDD276 pKa = 3.46LHH278 pKa = 6.68TLLQPSRR285 pKa = 11.84QVTFEE290 pKa = 3.71FDD292 pKa = 3.13NPAFSDD298 pKa = 3.65DD299 pKa = 3.63VSFEE303 pKa = 4.38FINDD307 pKa = 3.69LQEE310 pKa = 3.78VTAAPNPDD318 pKa = 3.37FQDD321 pKa = 2.95IQTLSRR327 pKa = 11.84PIISEE332 pKa = 4.17TEE334 pKa = 4.08SGSVRR339 pKa = 11.84YY340 pKa = 9.8SRR342 pKa = 11.84LGQRR346 pKa = 11.84ASMQTRR352 pKa = 11.84SGLVIGEE359 pKa = 4.33KK360 pKa = 9.73VHH362 pKa = 7.24FYY364 pKa = 11.22YY365 pKa = 10.66DD366 pKa = 3.72LSTIEE371 pKa = 4.29PADD374 pKa = 4.15SIEE377 pKa = 4.65LSTFVNTSADD387 pKa = 3.47AIVQNEE393 pKa = 4.14QIEE396 pKa = 4.71SLFIDD401 pKa = 4.09EE402 pKa = 5.55ASALLPDD409 pKa = 4.57EE410 pKa = 4.93EE411 pKa = 5.17LLDD414 pKa = 3.93SAAEE418 pKa = 4.16SFRR421 pKa = 11.84NSHH424 pKa = 6.33LVFSFTNEE432 pKa = 3.45EE433 pKa = 4.36AEE435 pKa = 4.55TVTLPILPSNFGLQPYY451 pKa = 10.19IPDD454 pKa = 3.33QFKK457 pKa = 10.87KK458 pKa = 10.57IFVSYY463 pKa = 9.26PISGSDD469 pKa = 3.41VAPYY473 pKa = 10.27LPSIPLSPLYY483 pKa = 8.52PTEE486 pKa = 3.82VTVYY490 pKa = 10.5GDD492 pKa = 3.77SFVIDD497 pKa = 3.66PFFLKK502 pKa = 10.34RR503 pKa = 11.84KK504 pKa = 8.0RR505 pKa = 11.84KK506 pKa = 9.91RR507 pKa = 11.84YY508 pKa = 7.13TLYY511 pKa = 11.02
MM1 pKa = 7.38SNNRR5 pKa = 11.84KK6 pKa = 9.02RR7 pKa = 11.84RR8 pKa = 11.84AAPIDD13 pKa = 4.13LYY15 pKa = 11.09KK16 pKa = 10.2SCQMGGDD23 pKa = 4.47CFPDD27 pKa = 3.35VQNKK31 pKa = 9.49IEE33 pKa = 4.19GTTLADD39 pKa = 3.55RR40 pKa = 11.84LLQIFSSVLYY50 pKa = 10.39FGGLGIGSGKK60 pKa = 8.44GTGGSTGYY68 pKa = 10.81KK69 pKa = 9.67PLTPGATQRR78 pKa = 11.84PSQFPQTFRR87 pKa = 11.84PTIPIDD93 pKa = 3.58PLGGADD99 pKa = 5.25VIPLDD104 pKa = 4.98IIDD107 pKa = 4.54ASTPSVVPLIDD118 pKa = 3.68GTPEE122 pKa = 3.63TTIISSGTGPTVEE135 pKa = 5.19IGDD138 pKa = 4.41LDD140 pKa = 3.68VTTTVDD146 pKa = 3.18ISATSVARR154 pKa = 11.84GDD156 pKa = 3.55HH157 pKa = 6.39PAIIDD162 pKa = 3.41ISEE165 pKa = 4.34EE166 pKa = 4.12GSSVIDD172 pKa = 3.88VQNGPPPPKK181 pKa = 10.08KK182 pKa = 10.4LQLDD186 pKa = 3.88TLLNPPEE193 pKa = 4.63TIDD196 pKa = 4.85LVTPPTTTYY205 pKa = 10.46RR206 pKa = 11.84DD207 pKa = 3.32SNINVFVDD215 pKa = 3.67THH217 pKa = 7.5ISGEE221 pKa = 4.34VIGAEE226 pKa = 4.29SIPLDD231 pKa = 3.68EE232 pKa = 5.69LSTIEE237 pKa = 3.84QFEE240 pKa = 4.29IEE242 pKa = 4.58EE243 pKa = 4.34PRR245 pKa = 11.84QTSTPEE251 pKa = 3.38ARR253 pKa = 11.84LNKK256 pKa = 10.07AIYY259 pKa = 8.93KK260 pKa = 10.57AKK262 pKa = 9.07TLYY265 pKa = 10.58NRR267 pKa = 11.84FVKK270 pKa = 10.0QVPTQDD276 pKa = 3.46LHH278 pKa = 6.68TLLQPSRR285 pKa = 11.84QVTFEE290 pKa = 3.71FDD292 pKa = 3.13NPAFSDD298 pKa = 3.65DD299 pKa = 3.63VSFEE303 pKa = 4.38FINDD307 pKa = 3.69LQEE310 pKa = 3.78VTAAPNPDD318 pKa = 3.37FQDD321 pKa = 2.95IQTLSRR327 pKa = 11.84PIISEE332 pKa = 4.17TEE334 pKa = 4.08SGSVRR339 pKa = 11.84YY340 pKa = 9.8SRR342 pKa = 11.84LGQRR346 pKa = 11.84ASMQTRR352 pKa = 11.84SGLVIGEE359 pKa = 4.33KK360 pKa = 9.73VHH362 pKa = 7.24FYY364 pKa = 11.22YY365 pKa = 10.66DD366 pKa = 3.72LSTIEE371 pKa = 4.29PADD374 pKa = 4.15SIEE377 pKa = 4.65LSTFVNTSADD387 pKa = 3.47AIVQNEE393 pKa = 4.14QIEE396 pKa = 4.71SLFIDD401 pKa = 4.09EE402 pKa = 5.55ASALLPDD409 pKa = 4.57EE410 pKa = 4.93EE411 pKa = 5.17LLDD414 pKa = 3.93SAAEE418 pKa = 4.16SFRR421 pKa = 11.84NSHH424 pKa = 6.33LVFSFTNEE432 pKa = 3.45EE433 pKa = 4.36AEE435 pKa = 4.55TVTLPILPSNFGLQPYY451 pKa = 10.19IPDD454 pKa = 3.33QFKK457 pKa = 10.87KK458 pKa = 10.57IFVSYY463 pKa = 9.26PISGSDD469 pKa = 3.41VAPYY473 pKa = 10.27LPSIPLSPLYY483 pKa = 8.52PTEE486 pKa = 3.82VTVYY490 pKa = 10.5GDD492 pKa = 3.77SFVIDD497 pKa = 3.66PFFLKK502 pKa = 10.34RR503 pKa = 11.84KK504 pKa = 8.0RR505 pKa = 11.84KK506 pKa = 9.91RR507 pKa = 11.84YY508 pKa = 7.13TLYY511 pKa = 11.02
Molecular weight: 55.98 kDa
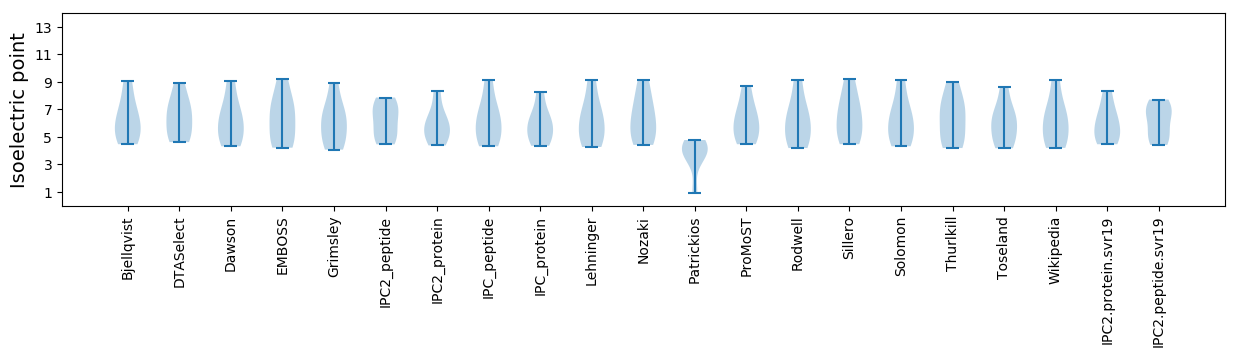
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8GMM8|G8GMM8_9PAPI E4 protein OS=human papillomavirus 153 OX=1110710 GN=E4 PE=4 SV=1
MM1 pKa = 6.96NQAEE5 pKa = 4.22LTARR9 pKa = 11.84FDD11 pKa = 3.78ALQEE15 pKa = 3.95EE16 pKa = 5.01LLNLIEE22 pKa = 4.85AAGDD26 pKa = 3.68DD27 pKa = 4.43LEE29 pKa = 4.39TQIRR33 pKa = 11.84HH34 pKa = 5.23WDD36 pKa = 3.56LNRR39 pKa = 11.84KK40 pKa = 9.19LNVYY44 pKa = 9.01MYY46 pKa = 9.43YY47 pKa = 10.64GRR49 pKa = 11.84KK50 pKa = 9.08EE51 pKa = 4.33GIRR54 pKa = 11.84HH55 pKa = 6.51FGLQQLPTLQVSEE68 pKa = 4.71YY69 pKa = 9.19NAKK72 pKa = 10.15LAISISILLKK82 pKa = 10.68SLAKK86 pKa = 10.45SRR88 pKa = 11.84FAYY91 pKa = 9.03EE92 pKa = 3.39KK93 pKa = 8.8WTFADD98 pKa = 4.21TSAEE102 pKa = 4.4LILTPPKK109 pKa = 10.73NCLKK113 pKa = 10.53KK114 pKa = 10.19SAYY117 pKa = 7.69TVQVIYY123 pKa = 10.91DD124 pKa = 4.33DD125 pKa = 5.38DD126 pKa = 4.51PEE128 pKa = 4.72NMSVHH133 pKa = 5.9TNWDD137 pKa = 3.43HH138 pKa = 7.86IYY140 pKa = 10.32YY141 pKa = 9.94QDD143 pKa = 6.09LEE145 pKa = 4.57EE146 pKa = 4.28QWHH149 pKa = 5.68KK150 pKa = 11.37VPGQVDD156 pKa = 4.13HH157 pKa = 7.31NGMFYY162 pKa = 11.12DD163 pKa = 5.21DD164 pKa = 3.89ILGEE168 pKa = 3.83RR169 pKa = 11.84VYY171 pKa = 10.67FHH173 pKa = 6.73VFAPDD178 pKa = 3.24ADD180 pKa = 4.37TYY182 pKa = 11.53SKK184 pKa = 10.02TGTWTVKK191 pKa = 10.59YY192 pKa = 10.03KK193 pKa = 9.87STTISSVVTSSSKK206 pKa = 9.92QSSRR210 pKa = 11.84GYY212 pKa = 9.06SARR215 pKa = 11.84DD216 pKa = 3.13STTSSRR222 pKa = 11.84CSSPEE227 pKa = 3.59EE228 pKa = 4.42GPSTRR233 pKa = 11.84RR234 pKa = 11.84SEE236 pKa = 4.16TSRR239 pKa = 11.84QEE241 pKa = 3.94SSGCSPTSSSLGNRR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84NQQGEE263 pKa = 4.55SEE265 pKa = 4.36SQRR268 pKa = 11.84DD269 pKa = 3.55SGGTSGKK276 pKa = 9.64RR277 pKa = 11.84KK278 pKa = 9.79RR279 pKa = 11.84PISLGVSAEE288 pKa = 4.26AVGSRR293 pKa = 11.84HH294 pKa = 6.33RR295 pKa = 11.84SLPRR299 pKa = 11.84QGLGRR304 pKa = 11.84LEE306 pKa = 4.25RR307 pKa = 11.84LTEE310 pKa = 3.92EE311 pKa = 4.84ARR313 pKa = 11.84DD314 pKa = 3.67PPIIIIKK321 pKa = 10.47GPANNLKK328 pKa = 10.1CWRR331 pKa = 11.84FRR333 pKa = 11.84CNNRR337 pKa = 11.84YY338 pKa = 8.71GHH340 pKa = 7.16LFSSISTVFRR350 pKa = 11.84WVEE353 pKa = 3.61DD354 pKa = 3.84CNNKK358 pKa = 9.74NEE360 pKa = 4.21AFFGSRR366 pKa = 11.84MLIAFKK372 pKa = 11.01DD373 pKa = 3.3NTQRR377 pKa = 11.84QKK379 pKa = 11.05FLSTVHH385 pKa = 6.79LPRR388 pKa = 11.84NTTYY392 pKa = 11.72ALGFLDD398 pKa = 4.7SLL400 pKa = 4.36
MM1 pKa = 6.96NQAEE5 pKa = 4.22LTARR9 pKa = 11.84FDD11 pKa = 3.78ALQEE15 pKa = 3.95EE16 pKa = 5.01LLNLIEE22 pKa = 4.85AAGDD26 pKa = 3.68DD27 pKa = 4.43LEE29 pKa = 4.39TQIRR33 pKa = 11.84HH34 pKa = 5.23WDD36 pKa = 3.56LNRR39 pKa = 11.84KK40 pKa = 9.19LNVYY44 pKa = 9.01MYY46 pKa = 9.43YY47 pKa = 10.64GRR49 pKa = 11.84KK50 pKa = 9.08EE51 pKa = 4.33GIRR54 pKa = 11.84HH55 pKa = 6.51FGLQQLPTLQVSEE68 pKa = 4.71YY69 pKa = 9.19NAKK72 pKa = 10.15LAISISILLKK82 pKa = 10.68SLAKK86 pKa = 10.45SRR88 pKa = 11.84FAYY91 pKa = 9.03EE92 pKa = 3.39KK93 pKa = 8.8WTFADD98 pKa = 4.21TSAEE102 pKa = 4.4LILTPPKK109 pKa = 10.73NCLKK113 pKa = 10.53KK114 pKa = 10.19SAYY117 pKa = 7.69TVQVIYY123 pKa = 10.91DD124 pKa = 4.33DD125 pKa = 5.38DD126 pKa = 4.51PEE128 pKa = 4.72NMSVHH133 pKa = 5.9TNWDD137 pKa = 3.43HH138 pKa = 7.86IYY140 pKa = 10.32YY141 pKa = 9.94QDD143 pKa = 6.09LEE145 pKa = 4.57EE146 pKa = 4.28QWHH149 pKa = 5.68KK150 pKa = 11.37VPGQVDD156 pKa = 4.13HH157 pKa = 7.31NGMFYY162 pKa = 11.12DD163 pKa = 5.21DD164 pKa = 3.89ILGEE168 pKa = 3.83RR169 pKa = 11.84VYY171 pKa = 10.67FHH173 pKa = 6.73VFAPDD178 pKa = 3.24ADD180 pKa = 4.37TYY182 pKa = 11.53SKK184 pKa = 10.02TGTWTVKK191 pKa = 10.59YY192 pKa = 10.03KK193 pKa = 9.87STTISSVVTSSSKK206 pKa = 9.92QSSRR210 pKa = 11.84GYY212 pKa = 9.06SARR215 pKa = 11.84DD216 pKa = 3.13STTSSRR222 pKa = 11.84CSSPEE227 pKa = 3.59EE228 pKa = 4.42GPSTRR233 pKa = 11.84RR234 pKa = 11.84SEE236 pKa = 4.16TSRR239 pKa = 11.84QEE241 pKa = 3.94SSGCSPTSSSLGNRR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84NQQGEE263 pKa = 4.55SEE265 pKa = 4.36SQRR268 pKa = 11.84DD269 pKa = 3.55SGGTSGKK276 pKa = 9.64RR277 pKa = 11.84KK278 pKa = 9.79RR279 pKa = 11.84PISLGVSAEE288 pKa = 4.26AVGSRR293 pKa = 11.84HH294 pKa = 6.33RR295 pKa = 11.84SLPRR299 pKa = 11.84QGLGRR304 pKa = 11.84LEE306 pKa = 4.25RR307 pKa = 11.84LTEE310 pKa = 3.92EE311 pKa = 4.84ARR313 pKa = 11.84DD314 pKa = 3.67PPIIIIKK321 pKa = 10.47GPANNLKK328 pKa = 10.1CWRR331 pKa = 11.84FRR333 pKa = 11.84CNNRR337 pKa = 11.84YY338 pKa = 8.71GHH340 pKa = 7.16LFSSISTVFRR350 pKa = 11.84WVEE353 pKa = 3.61DD354 pKa = 3.84CNNKK358 pKa = 9.74NEE360 pKa = 4.21AFFGSRR366 pKa = 11.84MLIAFKK372 pKa = 11.01DD373 pKa = 3.3NTQRR377 pKa = 11.84QKK379 pKa = 11.05FLSTVHH385 pKa = 6.79LPRR388 pKa = 11.84NTTYY392 pKa = 11.72ALGFLDD398 pKa = 4.7SLL400 pKa = 4.36
Molecular weight: 45.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2410 |
96 |
605 |
344.3 |
38.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.602 ± 0.531 | 2.365 ± 0.978 |
6.763 ± 0.445 | 6.1 ± 0.614 |
4.772 ± 0.429 | 5.187 ± 0.532 |
2.365 ± 0.556 | 5.726 ± 0.678 |
5.145 ± 0.63 | 9.585 ± 0.679 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.245 ± 0.25 | 5.145 ± 0.686 |
6.017 ± 1.113 | 4.564 ± 0.277 |
5.56 ± 0.713 | 7.718 ± 1.062 |
6.349 ± 0.804 | 5.394 ± 0.365 |
1.203 ± 0.356 | 3.195 ± 0.249 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
