
Hubei sobemo-like virus 26
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
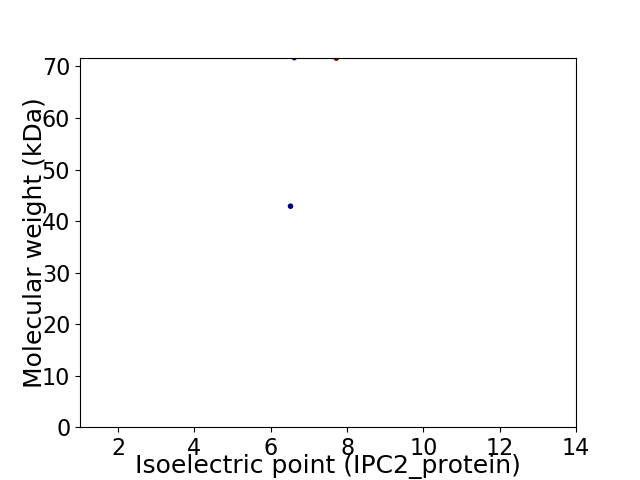
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE52|A0A1L3KE52_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 26 OX=1923212 PE=4 SV=1
MM1 pKa = 7.27RR2 pKa = 11.84QAVLDD7 pKa = 4.47ASCQAYY13 pKa = 8.97EE14 pKa = 3.94AARR17 pKa = 11.84WRR19 pKa = 11.84IPSDD23 pKa = 3.44FLQRR27 pKa = 11.84SHH29 pKa = 7.52FDD31 pKa = 2.86RR32 pKa = 11.84VVQNLDD38 pKa = 3.03WTSSPGYY45 pKa = 9.33PFLLRR50 pKa = 11.84APNNKK55 pKa = 9.07VLFGTDD61 pKa = 3.05DD62 pKa = 3.4QGCVDD67 pKa = 4.01EE68 pKa = 5.6AKK70 pKa = 10.81ASIIWQIVQDD80 pKa = 4.4RR81 pKa = 11.84ISGKK85 pKa = 10.04EE86 pKa = 3.35KK87 pKa = 10.31AGYY90 pKa = 9.05IRR92 pKa = 11.84LFIKK96 pKa = 10.71PEE98 pKa = 3.27AHH100 pKa = 5.77TAKK103 pKa = 10.56KK104 pKa = 10.32LADD107 pKa = 3.31EE108 pKa = 4.97RR109 pKa = 11.84YY110 pKa = 9.91RR111 pKa = 11.84LISSVNVVDD120 pKa = 4.94QIVDD124 pKa = 3.21HH125 pKa = 6.27MLFGDD130 pKa = 3.79MNGTCIANWPFIPSKK145 pKa = 10.08PGWSIAKK152 pKa = 9.8GGWRR156 pKa = 11.84FMPSEE161 pKa = 3.71VWMATDD167 pKa = 4.0ASAWDD172 pKa = 3.38WTVRR176 pKa = 11.84PWLLEE181 pKa = 3.69MALEE185 pKa = 4.9LRR187 pKa = 11.84MLLCDD192 pKa = 3.86NLSEE196 pKa = 4.22QWSTLATRR204 pKa = 11.84RR205 pKa = 11.84YY206 pKa = 9.94RR207 pKa = 11.84EE208 pKa = 4.73LFTQPEE214 pKa = 4.57FVTSGGLVLKK224 pKa = 10.57QKK226 pKa = 9.61TPGVMKK232 pKa = 10.13SGCVNTISDD241 pKa = 3.81NSIMQFILHH250 pKa = 6.36ARR252 pKa = 11.84VCLEE256 pKa = 4.13LGLPVTPMFSMGDD269 pKa = 3.39DD270 pKa = 3.2RR271 pKa = 11.84LQVPVRR277 pKa = 11.84DD278 pKa = 3.59QKK280 pKa = 11.68SYY282 pKa = 11.66VGLTSQFCILKK293 pKa = 10.23SVQLKK298 pKa = 10.71NEE300 pKa = 3.7FAGFEE305 pKa = 4.14FRR307 pKa = 11.84GSVVDD312 pKa = 4.13PVHH315 pKa = 6.61KK316 pKa = 10.42GKK318 pKa = 9.62HH319 pKa = 5.07AFNILHH325 pKa = 6.57MDD327 pKa = 3.87DD328 pKa = 4.3EE329 pKa = 4.97VAEE332 pKa = 4.46SMANSYY338 pKa = 9.32VLNYY342 pKa = 9.48HH343 pKa = 7.0RR344 pKa = 11.84SPWRR348 pKa = 11.84NWMEE352 pKa = 3.72RR353 pKa = 11.84LFNGMGIEE361 pKa = 4.7LIPRR365 pKa = 11.84TARR368 pKa = 11.84DD369 pKa = 3.57VIFDD373 pKa = 3.92GFF375 pKa = 3.56
MM1 pKa = 7.27RR2 pKa = 11.84QAVLDD7 pKa = 4.47ASCQAYY13 pKa = 8.97EE14 pKa = 3.94AARR17 pKa = 11.84WRR19 pKa = 11.84IPSDD23 pKa = 3.44FLQRR27 pKa = 11.84SHH29 pKa = 7.52FDD31 pKa = 2.86RR32 pKa = 11.84VVQNLDD38 pKa = 3.03WTSSPGYY45 pKa = 9.33PFLLRR50 pKa = 11.84APNNKK55 pKa = 9.07VLFGTDD61 pKa = 3.05DD62 pKa = 3.4QGCVDD67 pKa = 4.01EE68 pKa = 5.6AKK70 pKa = 10.81ASIIWQIVQDD80 pKa = 4.4RR81 pKa = 11.84ISGKK85 pKa = 10.04EE86 pKa = 3.35KK87 pKa = 10.31AGYY90 pKa = 9.05IRR92 pKa = 11.84LFIKK96 pKa = 10.71PEE98 pKa = 3.27AHH100 pKa = 5.77TAKK103 pKa = 10.56KK104 pKa = 10.32LADD107 pKa = 3.31EE108 pKa = 4.97RR109 pKa = 11.84YY110 pKa = 9.91RR111 pKa = 11.84LISSVNVVDD120 pKa = 4.94QIVDD124 pKa = 3.21HH125 pKa = 6.27MLFGDD130 pKa = 3.79MNGTCIANWPFIPSKK145 pKa = 10.08PGWSIAKK152 pKa = 9.8GGWRR156 pKa = 11.84FMPSEE161 pKa = 3.71VWMATDD167 pKa = 4.0ASAWDD172 pKa = 3.38WTVRR176 pKa = 11.84PWLLEE181 pKa = 3.69MALEE185 pKa = 4.9LRR187 pKa = 11.84MLLCDD192 pKa = 3.86NLSEE196 pKa = 4.22QWSTLATRR204 pKa = 11.84RR205 pKa = 11.84YY206 pKa = 9.94RR207 pKa = 11.84EE208 pKa = 4.73LFTQPEE214 pKa = 4.57FVTSGGLVLKK224 pKa = 10.57QKK226 pKa = 9.61TPGVMKK232 pKa = 10.13SGCVNTISDD241 pKa = 3.81NSIMQFILHH250 pKa = 6.36ARR252 pKa = 11.84VCLEE256 pKa = 4.13LGLPVTPMFSMGDD269 pKa = 3.39DD270 pKa = 3.2RR271 pKa = 11.84LQVPVRR277 pKa = 11.84DD278 pKa = 3.59QKK280 pKa = 11.68SYY282 pKa = 11.66VGLTSQFCILKK293 pKa = 10.23SVQLKK298 pKa = 10.71NEE300 pKa = 3.7FAGFEE305 pKa = 4.14FRR307 pKa = 11.84GSVVDD312 pKa = 4.13PVHH315 pKa = 6.61KK316 pKa = 10.42GKK318 pKa = 9.62HH319 pKa = 5.07AFNILHH325 pKa = 6.57MDD327 pKa = 3.87DD328 pKa = 4.3EE329 pKa = 4.97VAEE332 pKa = 4.46SMANSYY338 pKa = 9.32VLNYY342 pKa = 9.48HH343 pKa = 7.0RR344 pKa = 11.84SPWRR348 pKa = 11.84NWMEE352 pKa = 3.72RR353 pKa = 11.84LFNGMGIEE361 pKa = 4.7LIPRR365 pKa = 11.84TARR368 pKa = 11.84DD369 pKa = 3.57VIFDD373 pKa = 3.92GFF375 pKa = 3.56
Molecular weight: 42.88 kDa
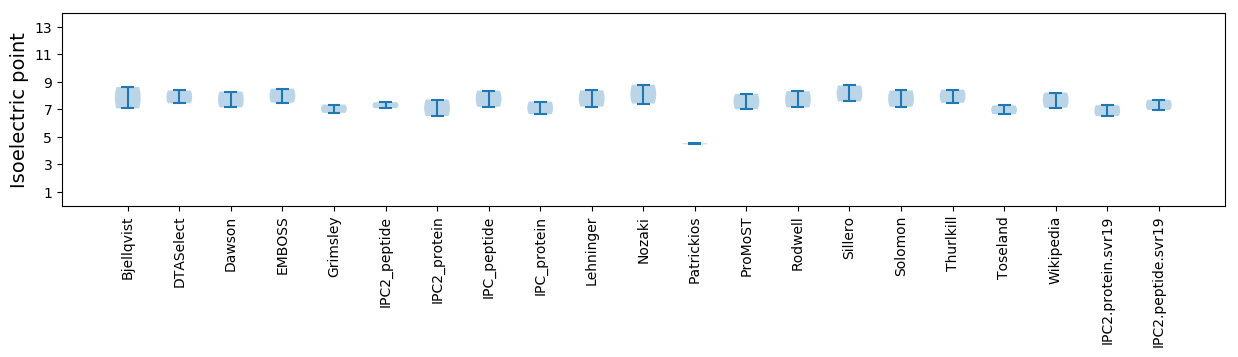
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE52|A0A1L3KE52_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 26 OX=1923212 PE=4 SV=1
MM1 pKa = 7.0NTTANTNNSEE11 pKa = 4.15GFQGEE16 pKa = 4.18PGGMFRR22 pKa = 11.84VRR24 pKa = 11.84LLGEE28 pKa = 4.12EE29 pKa = 4.03PVEE32 pKa = 3.79PVYY35 pKa = 10.79RR36 pKa = 11.84PTSEE40 pKa = 4.15EE41 pKa = 3.8VGEE44 pKa = 4.51SILTKK49 pKa = 10.49ALRR52 pKa = 11.84KK53 pKa = 9.26LAFWTGLMVLAALSIVSLLVIVYY76 pKa = 9.43RR77 pKa = 11.84WLAHH81 pKa = 5.79GAACVRR87 pKa = 11.84RR88 pKa = 11.84AVFSQFSYY96 pKa = 9.58MYY98 pKa = 9.43RR99 pKa = 11.84WLDD102 pKa = 3.49EE103 pKa = 4.3FSPSQTEE110 pKa = 3.79WEE112 pKa = 4.17RR113 pKa = 11.84AEE115 pKa = 4.03KK116 pKa = 10.51EE117 pKa = 3.67RR118 pKa = 11.84TLRR121 pKa = 11.84EE122 pKa = 3.7RR123 pKa = 11.84FGEE126 pKa = 4.13MWSQAVGFYY135 pKa = 10.52SVPLFLSGMDD145 pKa = 3.44WIDD148 pKa = 3.82WILLLAVVILTLIGLRR164 pKa = 11.84LAARR168 pKa = 11.84GCFSSGARR176 pKa = 11.84IVRR179 pKa = 11.84SWRR182 pKa = 11.84GVHH185 pKa = 5.36YY186 pKa = 10.24EE187 pKa = 3.59AMRR190 pKa = 11.84PGSFFVKK197 pKa = 10.36GDD199 pKa = 3.54VPDD202 pKa = 4.35CQVSVMLPGLLSDD215 pKa = 3.53SHH217 pKa = 7.34QGYY220 pKa = 9.42GIRR223 pKa = 11.84YY224 pKa = 8.21GEE226 pKa = 3.99YY227 pKa = 10.0LIVPRR232 pKa = 11.84HH233 pKa = 4.99VVAGHH238 pKa = 5.52QEE240 pKa = 4.45LILSSKK246 pKa = 8.64RR247 pKa = 11.84AKK249 pKa = 10.11VAVNVTFAQSRR260 pKa = 11.84LNEE263 pKa = 4.42DD264 pKa = 3.22LAYY267 pKa = 10.02MYY269 pKa = 10.55VGQNAFSQLGAVNAKK284 pKa = 9.72FPKK287 pKa = 10.38KK288 pKa = 10.43FMSTYY293 pKa = 8.69ATCTGLPGAVSARR306 pKa = 11.84VAKK309 pKa = 10.54SIIRR313 pKa = 11.84GKK315 pKa = 10.7LVFDD319 pKa = 4.83GSTLPGMSGAAYY331 pKa = 7.69MAQGLVLGIHH341 pKa = 5.94QGQSGMINVGLSADD355 pKa = 4.18LFRR358 pKa = 11.84AEE360 pKa = 3.49IKK362 pKa = 10.66YY363 pKa = 10.29LVRR366 pKa = 11.84QEE368 pKa = 4.59SVTGHH373 pKa = 6.1YY374 pKa = 10.68DD375 pKa = 3.05RR376 pKa = 11.84TDD378 pKa = 3.3DD379 pKa = 4.52DD380 pKa = 4.53RR381 pKa = 11.84PSKK384 pKa = 10.52FQANWNYY391 pKa = 10.8EE392 pKa = 4.36EE393 pKa = 4.18IEE395 pKa = 4.21KK396 pKa = 10.29MSEE399 pKa = 3.47LRR401 pKa = 11.84YY402 pKa = 10.44SQDD405 pKa = 2.46DD406 pKa = 3.67WAFDD410 pKa = 3.48AAIDD414 pKa = 4.22YY415 pKa = 9.97EE416 pKa = 4.25EE417 pKa = 4.2QLDD420 pKa = 3.99FGEE423 pKa = 4.54SMVRR427 pKa = 11.84KK428 pKa = 8.61PGPRR432 pKa = 11.84VRR434 pKa = 11.84LVPDD438 pKa = 3.05VEE440 pKa = 4.14GALRR444 pKa = 11.84VEE446 pKa = 4.53VAGQNTDD453 pKa = 3.08GKK455 pKa = 10.42SVSFDD460 pKa = 3.02AVQSHH465 pKa = 6.6HH466 pKa = 7.25LDD468 pKa = 3.28FLDD471 pKa = 3.47TLRR474 pKa = 11.84AANVLEE480 pKa = 4.63RR481 pKa = 11.84IEE483 pKa = 4.37ALEE486 pKa = 3.88SRR488 pKa = 11.84IKK490 pKa = 10.55PKK492 pKa = 10.53EE493 pKa = 3.52QVAPKK498 pKa = 9.94EE499 pKa = 4.18SAPPAFPCSEE509 pKa = 3.85CRR511 pKa = 11.84ARR513 pKa = 11.84CHH515 pKa = 6.1TEE517 pKa = 3.61EE518 pKa = 4.15KK519 pKa = 10.24LANHH523 pKa = 5.73MMVHH527 pKa = 5.72KK528 pKa = 10.25QYY530 pKa = 10.72PCEE533 pKa = 4.25SCDD536 pKa = 3.83KK537 pKa = 10.9VFTGEE542 pKa = 3.85AKK544 pKa = 10.63LARR547 pKa = 11.84HH548 pKa = 6.2LRR550 pKa = 11.84SAHH553 pKa = 5.61PVQPEE558 pKa = 4.05SAVPEE563 pKa = 4.15DD564 pKa = 3.44TGKK567 pKa = 10.45SGRR570 pKa = 11.84QVKK573 pKa = 8.35MEE575 pKa = 4.17KK576 pKa = 10.56VPFLGKK582 pKa = 10.09KK583 pKa = 10.15ASNQNRR589 pKa = 11.84KK590 pKa = 9.67KK591 pKa = 10.68SLSLNSSASAGKK603 pKa = 10.01KK604 pKa = 9.68GSPSVEE610 pKa = 3.52ASQCLISEE618 pKa = 4.54FQKK621 pKa = 11.14SMRR624 pKa = 11.84VLLKK628 pKa = 10.7EE629 pKa = 4.02FAEE632 pKa = 4.44TLAGQNSATMRR643 pKa = 11.84NN644 pKa = 3.63
MM1 pKa = 7.0NTTANTNNSEE11 pKa = 4.15GFQGEE16 pKa = 4.18PGGMFRR22 pKa = 11.84VRR24 pKa = 11.84LLGEE28 pKa = 4.12EE29 pKa = 4.03PVEE32 pKa = 3.79PVYY35 pKa = 10.79RR36 pKa = 11.84PTSEE40 pKa = 4.15EE41 pKa = 3.8VGEE44 pKa = 4.51SILTKK49 pKa = 10.49ALRR52 pKa = 11.84KK53 pKa = 9.26LAFWTGLMVLAALSIVSLLVIVYY76 pKa = 9.43RR77 pKa = 11.84WLAHH81 pKa = 5.79GAACVRR87 pKa = 11.84RR88 pKa = 11.84AVFSQFSYY96 pKa = 9.58MYY98 pKa = 9.43RR99 pKa = 11.84WLDD102 pKa = 3.49EE103 pKa = 4.3FSPSQTEE110 pKa = 3.79WEE112 pKa = 4.17RR113 pKa = 11.84AEE115 pKa = 4.03KK116 pKa = 10.51EE117 pKa = 3.67RR118 pKa = 11.84TLRR121 pKa = 11.84EE122 pKa = 3.7RR123 pKa = 11.84FGEE126 pKa = 4.13MWSQAVGFYY135 pKa = 10.52SVPLFLSGMDD145 pKa = 3.44WIDD148 pKa = 3.82WILLLAVVILTLIGLRR164 pKa = 11.84LAARR168 pKa = 11.84GCFSSGARR176 pKa = 11.84IVRR179 pKa = 11.84SWRR182 pKa = 11.84GVHH185 pKa = 5.36YY186 pKa = 10.24EE187 pKa = 3.59AMRR190 pKa = 11.84PGSFFVKK197 pKa = 10.36GDD199 pKa = 3.54VPDD202 pKa = 4.35CQVSVMLPGLLSDD215 pKa = 3.53SHH217 pKa = 7.34QGYY220 pKa = 9.42GIRR223 pKa = 11.84YY224 pKa = 8.21GEE226 pKa = 3.99YY227 pKa = 10.0LIVPRR232 pKa = 11.84HH233 pKa = 4.99VVAGHH238 pKa = 5.52QEE240 pKa = 4.45LILSSKK246 pKa = 8.64RR247 pKa = 11.84AKK249 pKa = 10.11VAVNVTFAQSRR260 pKa = 11.84LNEE263 pKa = 4.42DD264 pKa = 3.22LAYY267 pKa = 10.02MYY269 pKa = 10.55VGQNAFSQLGAVNAKK284 pKa = 9.72FPKK287 pKa = 10.38KK288 pKa = 10.43FMSTYY293 pKa = 8.69ATCTGLPGAVSARR306 pKa = 11.84VAKK309 pKa = 10.54SIIRR313 pKa = 11.84GKK315 pKa = 10.7LVFDD319 pKa = 4.83GSTLPGMSGAAYY331 pKa = 7.69MAQGLVLGIHH341 pKa = 5.94QGQSGMINVGLSADD355 pKa = 4.18LFRR358 pKa = 11.84AEE360 pKa = 3.49IKK362 pKa = 10.66YY363 pKa = 10.29LVRR366 pKa = 11.84QEE368 pKa = 4.59SVTGHH373 pKa = 6.1YY374 pKa = 10.68DD375 pKa = 3.05RR376 pKa = 11.84TDD378 pKa = 3.3DD379 pKa = 4.52DD380 pKa = 4.53RR381 pKa = 11.84PSKK384 pKa = 10.52FQANWNYY391 pKa = 10.8EE392 pKa = 4.36EE393 pKa = 4.18IEE395 pKa = 4.21KK396 pKa = 10.29MSEE399 pKa = 3.47LRR401 pKa = 11.84YY402 pKa = 10.44SQDD405 pKa = 2.46DD406 pKa = 3.67WAFDD410 pKa = 3.48AAIDD414 pKa = 4.22YY415 pKa = 9.97EE416 pKa = 4.25EE417 pKa = 4.2QLDD420 pKa = 3.99FGEE423 pKa = 4.54SMVRR427 pKa = 11.84KK428 pKa = 8.61PGPRR432 pKa = 11.84VRR434 pKa = 11.84LVPDD438 pKa = 3.05VEE440 pKa = 4.14GALRR444 pKa = 11.84VEE446 pKa = 4.53VAGQNTDD453 pKa = 3.08GKK455 pKa = 10.42SVSFDD460 pKa = 3.02AVQSHH465 pKa = 6.6HH466 pKa = 7.25LDD468 pKa = 3.28FLDD471 pKa = 3.47TLRR474 pKa = 11.84AANVLEE480 pKa = 4.63RR481 pKa = 11.84IEE483 pKa = 4.37ALEE486 pKa = 3.88SRR488 pKa = 11.84IKK490 pKa = 10.55PKK492 pKa = 10.53EE493 pKa = 3.52QVAPKK498 pKa = 9.94EE499 pKa = 4.18SAPPAFPCSEE509 pKa = 3.85CRR511 pKa = 11.84ARR513 pKa = 11.84CHH515 pKa = 6.1TEE517 pKa = 3.61EE518 pKa = 4.15KK519 pKa = 10.24LANHH523 pKa = 5.73MMVHH527 pKa = 5.72KK528 pKa = 10.25QYY530 pKa = 10.72PCEE533 pKa = 4.25SCDD536 pKa = 3.83KK537 pKa = 10.9VFTGEE542 pKa = 3.85AKK544 pKa = 10.63LARR547 pKa = 11.84HH548 pKa = 6.2LRR550 pKa = 11.84SAHH553 pKa = 5.61PVQPEE558 pKa = 4.05SAVPEE563 pKa = 4.15DD564 pKa = 3.44TGKK567 pKa = 10.45SGRR570 pKa = 11.84QVKK573 pKa = 8.35MEE575 pKa = 4.17KK576 pKa = 10.56VPFLGKK582 pKa = 10.09KK583 pKa = 10.15ASNQNRR589 pKa = 11.84KK590 pKa = 9.67KK591 pKa = 10.68SLSLNSSASAGKK603 pKa = 10.01KK604 pKa = 9.68GSPSVEE610 pKa = 3.52ASQCLISEE618 pKa = 4.54FQKK621 pKa = 11.14SMRR624 pKa = 11.84VLLKK628 pKa = 10.7EE629 pKa = 4.02FAEE632 pKa = 4.44TLAGQNSATMRR643 pKa = 11.84NN644 pKa = 3.63
Molecular weight: 71.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1019 |
375 |
644 |
509.5 |
57.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.243 ± 0.831 | 1.668 ± 0.105 |
4.809 ± 0.839 | 6.379 ± 0.832 |
4.711 ± 0.328 | 6.968 ± 0.44 |
2.159 ± 0.014 | 4.024 ± 0.69 |
5.005 ± 0.249 | 8.93 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.435 ± 0.298 | 3.337 ± 0.35 |
4.514 ± 0.151 | 4.22 ± 0.025 |
6.771 ± 0.055 | 8.243 ± 0.55 |
3.729 ± 0.143 | 7.949 ± 0.254 |
2.257 ± 0.637 | 2.65 ± 0.272 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
