
Brachybacterium muris UCD-AY4
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Dermabacteraceae; Brachybacterium; Brachybacterium muris
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
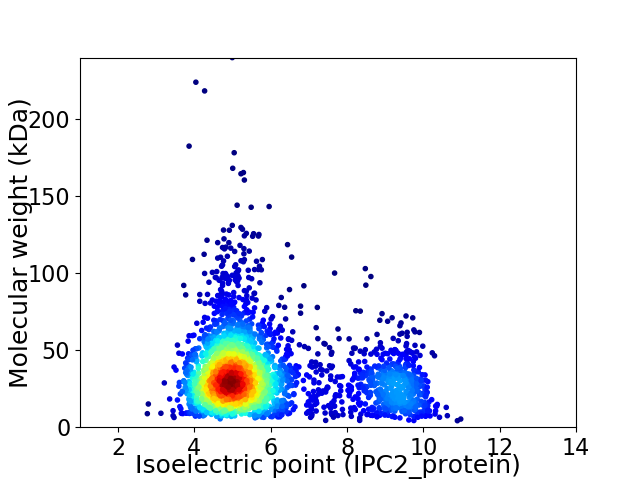
Virtual 2D-PAGE plot for 2806 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A022L0R3|A0A022L0R3_9MICO Amidophosphoribosyltransferase OS=Brachybacterium muris UCD-AY4 OX=1249481 GN=purF PE=3 SV=1
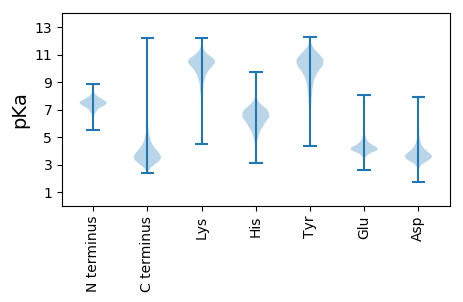
MM1 pKa = 7.91RR2 pKa = 11.84LGSTPRR8 pKa = 11.84RR9 pKa = 11.84SPAHH13 pKa = 6.35RR14 pKa = 11.84PPTRR18 pKa = 11.84RR19 pKa = 11.84PLARR23 pKa = 11.84LSAVAAAAVLVVAGCVPQEE42 pKa = 4.01GQGPGEE48 pKa = 4.3TPSTGQSDD56 pKa = 3.44NGGAGSGDD64 pKa = 3.37GTTDD68 pKa = 3.07GTSGDD73 pKa = 3.95GQEE76 pKa = 4.43LATLGTSAAKK86 pKa = 10.16DD87 pKa = 3.83LPTDD91 pKa = 3.72PAEE94 pKa = 4.61DD95 pKa = 3.46PAYY98 pKa = 10.32AAYY101 pKa = 10.42YY102 pKa = 9.0EE103 pKa = 4.54QDD105 pKa = 3.77ITWGPCDD112 pKa = 3.77DD113 pKa = 4.12VEE115 pKa = 5.26GGSEE119 pKa = 4.37LEE121 pKa = 4.64CGTLTVPMVWNDD133 pKa = 3.65PGAGDD138 pKa = 3.82IDD140 pKa = 4.3LAVARR145 pKa = 11.84HH146 pKa = 5.36AASGNRR152 pKa = 11.84TGSLILNPGGPGGSGVDD169 pKa = 4.42FTASAPFLFSRR180 pKa = 11.84PVVDD184 pKa = 5.06AYY186 pKa = 10.93DD187 pKa = 3.8IIGFDD192 pKa = 3.47PRR194 pKa = 11.84GVARR198 pKa = 11.84SAGIQCLSDD207 pKa = 3.72EE208 pKa = 4.48EE209 pKa = 4.31TDD211 pKa = 3.48EE212 pKa = 4.23YY213 pKa = 11.54LAGTGDD219 pKa = 4.1MGTEE223 pKa = 4.06DD224 pKa = 4.18PLEE227 pKa = 4.5EE228 pKa = 4.0GLAWMKK234 pKa = 10.63RR235 pKa = 11.84IAEE238 pKa = 4.46GCQQDD243 pKa = 3.62SGEE246 pKa = 4.42LLPYY250 pKa = 10.41LDD252 pKa = 4.52TYY254 pKa = 11.15SAARR258 pKa = 11.84DD259 pKa = 3.61MDD261 pKa = 4.06VLRR264 pKa = 11.84AAVDD268 pKa = 3.88SEE270 pKa = 3.72QLDD273 pKa = 4.0YY274 pKa = 11.21FGYY277 pKa = 10.47SYY279 pKa = 10.34GTYY282 pKa = 10.46LGATYY287 pKa = 11.09ADD289 pKa = 5.2LYY291 pKa = 9.42PEE293 pKa = 4.15RR294 pKa = 11.84VGKK297 pKa = 9.92FVLDD301 pKa = 3.77AALDD305 pKa = 3.93PSSTLDD311 pKa = 3.85EE312 pKa = 4.56ISAGQAEE319 pKa = 4.79GFEE322 pKa = 4.4LATDD326 pKa = 4.52AFLEE330 pKa = 4.52DD331 pKa = 4.97CLAQGDD337 pKa = 4.18SCPFKK342 pKa = 11.21GEE344 pKa = 3.83PAEE347 pKa = 5.56ANQQLQNFFDD357 pKa = 4.47SIEE360 pKa = 4.16AQPLEE365 pKa = 4.31TSDD368 pKa = 3.49PQRR371 pKa = 11.84PLTVSLAQSAVLLLMYY387 pKa = 10.38EE388 pKa = 4.27DD389 pKa = 5.17GLWQVGRR396 pKa = 11.84DD397 pKa = 3.53ALAQGMQGDD406 pKa = 5.06GSALLQIADD415 pKa = 4.5LSSEE419 pKa = 4.09RR420 pKa = 11.84QPDD423 pKa = 3.55GSYY426 pKa = 10.45KK427 pKa = 10.23GNGMFAINAINCLDD441 pKa = 3.75HH442 pKa = 7.39PGIADD447 pKa = 3.68QEE449 pKa = 4.42WQAKK453 pKa = 7.11EE454 pKa = 4.06AEE456 pKa = 4.3RR457 pKa = 11.84LAAEE461 pKa = 4.48YY462 pKa = 7.54PTFGPSMGYY471 pKa = 9.89GATMCAQWPVPPLRR485 pKa = 11.84EE486 pKa = 3.95PAPISAEE493 pKa = 3.85GAGPIVVIGTTGDD506 pKa = 3.32PATPYY511 pKa = 10.19RR512 pKa = 11.84WSQSLAEE519 pKa = 4.14QLDD522 pKa = 4.0DD523 pKa = 4.8AVLLTFEE530 pKa = 5.2GNGHH534 pKa = 4.96TAYY537 pKa = 10.14GRR539 pKa = 11.84SGGCIEE545 pKa = 4.53EE546 pKa = 4.85AVDD549 pKa = 4.27AYY551 pKa = 10.98LLEE554 pKa = 4.65GTTPQDD560 pKa = 3.55GLTCSSGGG568 pKa = 3.46
MM1 pKa = 7.91RR2 pKa = 11.84LGSTPRR8 pKa = 11.84RR9 pKa = 11.84SPAHH13 pKa = 6.35RR14 pKa = 11.84PPTRR18 pKa = 11.84RR19 pKa = 11.84PLARR23 pKa = 11.84LSAVAAAAVLVVAGCVPQEE42 pKa = 4.01GQGPGEE48 pKa = 4.3TPSTGQSDD56 pKa = 3.44NGGAGSGDD64 pKa = 3.37GTTDD68 pKa = 3.07GTSGDD73 pKa = 3.95GQEE76 pKa = 4.43LATLGTSAAKK86 pKa = 10.16DD87 pKa = 3.83LPTDD91 pKa = 3.72PAEE94 pKa = 4.61DD95 pKa = 3.46PAYY98 pKa = 10.32AAYY101 pKa = 10.42YY102 pKa = 9.0EE103 pKa = 4.54QDD105 pKa = 3.77ITWGPCDD112 pKa = 3.77DD113 pKa = 4.12VEE115 pKa = 5.26GGSEE119 pKa = 4.37LEE121 pKa = 4.64CGTLTVPMVWNDD133 pKa = 3.65PGAGDD138 pKa = 3.82IDD140 pKa = 4.3LAVARR145 pKa = 11.84HH146 pKa = 5.36AASGNRR152 pKa = 11.84TGSLILNPGGPGGSGVDD169 pKa = 4.42FTASAPFLFSRR180 pKa = 11.84PVVDD184 pKa = 5.06AYY186 pKa = 10.93DD187 pKa = 3.8IIGFDD192 pKa = 3.47PRR194 pKa = 11.84GVARR198 pKa = 11.84SAGIQCLSDD207 pKa = 3.72EE208 pKa = 4.48EE209 pKa = 4.31TDD211 pKa = 3.48EE212 pKa = 4.23YY213 pKa = 11.54LAGTGDD219 pKa = 4.1MGTEE223 pKa = 4.06DD224 pKa = 4.18PLEE227 pKa = 4.5EE228 pKa = 4.0GLAWMKK234 pKa = 10.63RR235 pKa = 11.84IAEE238 pKa = 4.46GCQQDD243 pKa = 3.62SGEE246 pKa = 4.42LLPYY250 pKa = 10.41LDD252 pKa = 4.52TYY254 pKa = 11.15SAARR258 pKa = 11.84DD259 pKa = 3.61MDD261 pKa = 4.06VLRR264 pKa = 11.84AAVDD268 pKa = 3.88SEE270 pKa = 3.72QLDD273 pKa = 4.0YY274 pKa = 11.21FGYY277 pKa = 10.47SYY279 pKa = 10.34GTYY282 pKa = 10.46LGATYY287 pKa = 11.09ADD289 pKa = 5.2LYY291 pKa = 9.42PEE293 pKa = 4.15RR294 pKa = 11.84VGKK297 pKa = 9.92FVLDD301 pKa = 3.77AALDD305 pKa = 3.93PSSTLDD311 pKa = 3.85EE312 pKa = 4.56ISAGQAEE319 pKa = 4.79GFEE322 pKa = 4.4LATDD326 pKa = 4.52AFLEE330 pKa = 4.52DD331 pKa = 4.97CLAQGDD337 pKa = 4.18SCPFKK342 pKa = 11.21GEE344 pKa = 3.83PAEE347 pKa = 5.56ANQQLQNFFDD357 pKa = 4.47SIEE360 pKa = 4.16AQPLEE365 pKa = 4.31TSDD368 pKa = 3.49PQRR371 pKa = 11.84PLTVSLAQSAVLLLMYY387 pKa = 10.38EE388 pKa = 4.27DD389 pKa = 5.17GLWQVGRR396 pKa = 11.84DD397 pKa = 3.53ALAQGMQGDD406 pKa = 5.06GSALLQIADD415 pKa = 4.5LSSEE419 pKa = 4.09RR420 pKa = 11.84QPDD423 pKa = 3.55GSYY426 pKa = 10.45KK427 pKa = 10.23GNGMFAINAINCLDD441 pKa = 3.75HH442 pKa = 7.39PGIADD447 pKa = 3.68QEE449 pKa = 4.42WQAKK453 pKa = 7.11EE454 pKa = 4.06AEE456 pKa = 4.3RR457 pKa = 11.84LAAEE461 pKa = 4.48YY462 pKa = 7.54PTFGPSMGYY471 pKa = 9.89GATMCAQWPVPPLRR485 pKa = 11.84EE486 pKa = 3.95PAPISAEE493 pKa = 3.85GAGPIVVIGTTGDD506 pKa = 3.32PATPYY511 pKa = 10.19RR512 pKa = 11.84WSQSLAEE519 pKa = 4.14QLDD522 pKa = 4.0DD523 pKa = 4.8AVLLTFEE530 pKa = 5.2GNGHH534 pKa = 4.96TAYY537 pKa = 10.14GRR539 pKa = 11.84SGGCIEE545 pKa = 4.53EE546 pKa = 4.85AVDD549 pKa = 4.27AYY551 pKa = 10.98LLEE554 pKa = 4.65GTTPQDD560 pKa = 3.55GLTCSSGGG568 pKa = 3.46
Molecular weight: 59.39 kDa
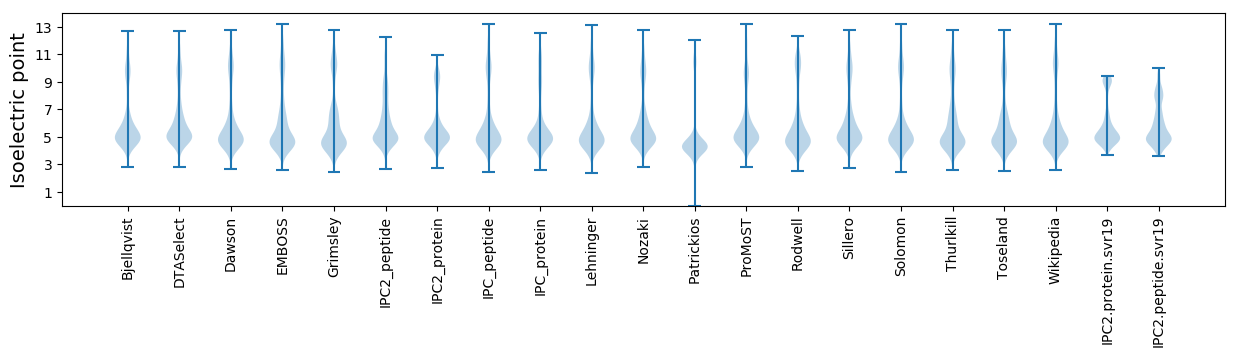
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A022L020|A0A022L020_9MICO Uncharacterized protein OS=Brachybacterium muris UCD-AY4 OX=1249481 GN=D641_0109990 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIINARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84SEE42 pKa = 4.37LSAA45 pKa = 4.77
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIINARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84SEE42 pKa = 4.37LSAA45 pKa = 4.77
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
943634 |
37 |
2201 |
336.3 |
36.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.726 ± 0.06 | 0.558 ± 0.011 |
6.393 ± 0.04 | 6.283 ± 0.038 |
2.686 ± 0.03 | 9.121 ± 0.043 |
2.27 ± 0.024 | 4.129 ± 0.035 |
1.869 ± 0.032 | 10.271 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.027 ± 0.018 | 1.7 ± 0.024 |
5.799 ± 0.035 | 3.218 ± 0.023 |
7.578 ± 0.054 | 5.529 ± 0.028 |
6.22 ± 0.033 | 8.447 ± 0.048 |
1.387 ± 0.019 | 1.788 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
