
Bat polyomavirus 6c
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus; Dobsonia moluccensis polyomavirus 3
Average proteome isoelectric point is 5.62
Get precalculated fractions of proteins
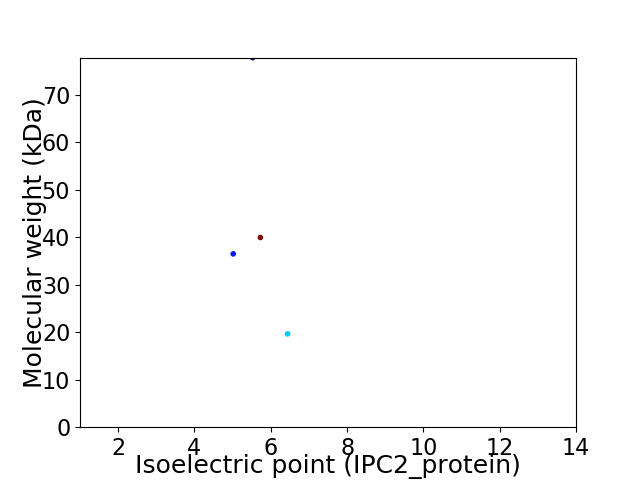
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
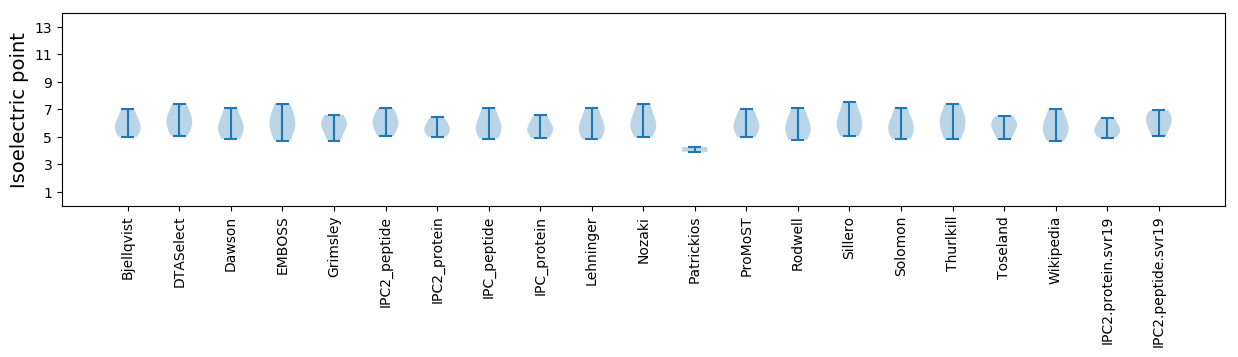
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D5ZYM9|A0A0D5ZYM9_9POLY Small t antigen OS=Bat polyomavirus 6c OX=1623688 PE=4 SV=1
MM1 pKa = 7.39GALLAVIAEE10 pKa = 4.31VAEE13 pKa = 4.37VSAVSGLSVDD23 pKa = 3.86ALLSGEE29 pKa = 4.27ALASAEE35 pKa = 4.28LIAGQVEE42 pKa = 4.86AISALEE48 pKa = 3.89GLSAVEE54 pKa = 4.08AAATLGFSAEE64 pKa = 4.18NYY66 pKa = 9.49NAIAALASLPNALSGIVGADD86 pKa = 3.11LVFNGTSVVAAAISTAVSPYY106 pKa = 9.01TYY108 pKa = 9.78DD109 pKa = 3.31YY110 pKa = 11.24SVPITNLNVNMALQVWQPDD129 pKa = 3.63LDD131 pKa = 4.09DD132 pKa = 5.53LNFPGVMPFVRR143 pKa = 11.84FLNYY147 pKa = 9.14IDD149 pKa = 3.9PMHH152 pKa = 6.68WAGGLFRR159 pKa = 11.84ALGRR163 pKa = 11.84YY164 pKa = 7.6FWQNAQRR171 pKa = 11.84QGQQMLEE178 pKa = 4.09GEE180 pKa = 4.49FRR182 pKa = 11.84EE183 pKa = 4.27LAQRR187 pKa = 11.84TATSISEE194 pKa = 4.01TLALYY199 pKa = 10.07FEE201 pKa = 4.41NARR204 pKa = 11.84WAVRR208 pKa = 11.84AVPTEE213 pKa = 3.81LYY215 pKa = 10.91GRR217 pKa = 11.84LQQYY221 pKa = 8.81YY222 pKa = 10.89AEE224 pKa = 5.08LPPLNPPQLRR234 pKa = 11.84EE235 pKa = 3.9VSRR238 pKa = 11.84RR239 pKa = 11.84TANQPFQLYY248 pKa = 10.73DD249 pKa = 4.08KK250 pKa = 10.14ILSNVEE256 pKa = 3.58SAHH259 pKa = 5.88YY260 pKa = 7.82VTKK263 pKa = 10.58VDD265 pKa = 3.97PPGGANQRR273 pKa = 11.84QTPDD277 pKa = 2.7WLLPLILGLYY287 pKa = 10.37GDD289 pKa = 5.19ISPSWEE295 pKa = 3.98TTLEE299 pKa = 3.98EE300 pKa = 5.04LEE302 pKa = 4.4EE303 pKa = 4.52EE304 pKa = 4.01EE305 pKa = 5.96DD306 pKa = 4.01GPKK309 pKa = 10.07KK310 pKa = 10.38KK311 pKa = 10.12KK312 pKa = 9.91QRR314 pKa = 11.84PSTKK318 pKa = 7.82TTPHH322 pKa = 6.98RR323 pKa = 11.84SSKK326 pKa = 8.8TYY328 pKa = 9.97SKK330 pKa = 10.44RR331 pKa = 11.84RR332 pKa = 11.84HH333 pKa = 5.04
MM1 pKa = 7.39GALLAVIAEE10 pKa = 4.31VAEE13 pKa = 4.37VSAVSGLSVDD23 pKa = 3.86ALLSGEE29 pKa = 4.27ALASAEE35 pKa = 4.28LIAGQVEE42 pKa = 4.86AISALEE48 pKa = 3.89GLSAVEE54 pKa = 4.08AAATLGFSAEE64 pKa = 4.18NYY66 pKa = 9.49NAIAALASLPNALSGIVGADD86 pKa = 3.11LVFNGTSVVAAAISTAVSPYY106 pKa = 9.01TYY108 pKa = 9.78DD109 pKa = 3.31YY110 pKa = 11.24SVPITNLNVNMALQVWQPDD129 pKa = 3.63LDD131 pKa = 4.09DD132 pKa = 5.53LNFPGVMPFVRR143 pKa = 11.84FLNYY147 pKa = 9.14IDD149 pKa = 3.9PMHH152 pKa = 6.68WAGGLFRR159 pKa = 11.84ALGRR163 pKa = 11.84YY164 pKa = 7.6FWQNAQRR171 pKa = 11.84QGQQMLEE178 pKa = 4.09GEE180 pKa = 4.49FRR182 pKa = 11.84EE183 pKa = 4.27LAQRR187 pKa = 11.84TATSISEE194 pKa = 4.01TLALYY199 pKa = 10.07FEE201 pKa = 4.41NARR204 pKa = 11.84WAVRR208 pKa = 11.84AVPTEE213 pKa = 3.81LYY215 pKa = 10.91GRR217 pKa = 11.84LQQYY221 pKa = 8.81YY222 pKa = 10.89AEE224 pKa = 5.08LPPLNPPQLRR234 pKa = 11.84EE235 pKa = 3.9VSRR238 pKa = 11.84RR239 pKa = 11.84TANQPFQLYY248 pKa = 10.73DD249 pKa = 4.08KK250 pKa = 10.14ILSNVEE256 pKa = 3.58SAHH259 pKa = 5.88YY260 pKa = 7.82VTKK263 pKa = 10.58VDD265 pKa = 3.97PPGGANQRR273 pKa = 11.84QTPDD277 pKa = 2.7WLLPLILGLYY287 pKa = 10.37GDD289 pKa = 5.19ISPSWEE295 pKa = 3.98TTLEE299 pKa = 3.98EE300 pKa = 5.04LEE302 pKa = 4.4EE303 pKa = 4.52EE304 pKa = 4.01EE305 pKa = 5.96DD306 pKa = 4.01GPKK309 pKa = 10.07KK310 pKa = 10.38KK311 pKa = 10.12KK312 pKa = 9.91QRR314 pKa = 11.84PSTKK318 pKa = 7.82TTPHH322 pKa = 6.98RR323 pKa = 11.84SSKK326 pKa = 8.8TYY328 pKa = 9.97SKK330 pKa = 10.44RR331 pKa = 11.84RR332 pKa = 11.84HH333 pKa = 5.04
Molecular weight: 36.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D5ZYR4|A0A0D5ZYR4_9POLY Capsid protein VP1 OS=Bat polyomavirus 6c OX=1623688 GN=VP1 PE=3 SV=1
MM1 pKa = 7.66DD2 pKa = 4.87HH3 pKa = 7.31ALTRR7 pKa = 11.84EE8 pKa = 3.82EE9 pKa = 4.2SQRR12 pKa = 11.84LMEE15 pKa = 4.87LLNLPMEE22 pKa = 4.43QFGNFHH28 pKa = 5.99LMKK31 pKa = 10.33RR32 pKa = 11.84AFLSSIKK39 pKa = 10.08KK40 pKa = 7.26YY41 pKa = 10.73HH42 pKa = 6.79PDD44 pKa = 3.29KK45 pKa = 11.59GGDD48 pKa = 3.01EE49 pKa = 4.46SMAKK53 pKa = 10.08EE54 pKa = 4.83LISLYY59 pKa = 10.58KK60 pKa = 10.29KK61 pKa = 10.68AEE63 pKa = 4.1CHH65 pKa = 5.45VSSLEE70 pKa = 4.22TEE72 pKa = 4.6DD73 pKa = 3.89DD74 pKa = 3.76TSFTTDD80 pKa = 3.05EE81 pKa = 4.13VQKK84 pKa = 11.48ADD86 pKa = 3.27MFLYY90 pKa = 10.5LRR92 pKa = 11.84DD93 pKa = 3.49WVEE96 pKa = 4.47CNMGFPCKK104 pKa = 10.22CLFCMLRR111 pKa = 11.84KK112 pKa = 9.5RR113 pKa = 11.84HH114 pKa = 5.62NEE116 pKa = 3.55RR117 pKa = 11.84KK118 pKa = 9.94KK119 pKa = 10.82NLLHH123 pKa = 6.32NVWGTCYY130 pKa = 10.35CFKK133 pKa = 10.85CYY135 pKa = 10.1ILWFGLEE142 pKa = 4.02HH143 pKa = 6.21SWIIFLSWKK152 pKa = 10.33GIIANTPYY160 pKa = 10.65RR161 pKa = 11.84CLDD164 pKa = 3.47LL165 pKa = 6.4
MM1 pKa = 7.66DD2 pKa = 4.87HH3 pKa = 7.31ALTRR7 pKa = 11.84EE8 pKa = 3.82EE9 pKa = 4.2SQRR12 pKa = 11.84LMEE15 pKa = 4.87LLNLPMEE22 pKa = 4.43QFGNFHH28 pKa = 5.99LMKK31 pKa = 10.33RR32 pKa = 11.84AFLSSIKK39 pKa = 10.08KK40 pKa = 7.26YY41 pKa = 10.73HH42 pKa = 6.79PDD44 pKa = 3.29KK45 pKa = 11.59GGDD48 pKa = 3.01EE49 pKa = 4.46SMAKK53 pKa = 10.08EE54 pKa = 4.83LISLYY59 pKa = 10.58KK60 pKa = 10.29KK61 pKa = 10.68AEE63 pKa = 4.1CHH65 pKa = 5.45VSSLEE70 pKa = 4.22TEE72 pKa = 4.6DD73 pKa = 3.89DD74 pKa = 3.76TSFTTDD80 pKa = 3.05EE81 pKa = 4.13VQKK84 pKa = 11.48ADD86 pKa = 3.27MFLYY90 pKa = 10.5LRR92 pKa = 11.84DD93 pKa = 3.49WVEE96 pKa = 4.47CNMGFPCKK104 pKa = 10.22CLFCMLRR111 pKa = 11.84KK112 pKa = 9.5RR113 pKa = 11.84HH114 pKa = 5.62NEE116 pKa = 3.55RR117 pKa = 11.84KK118 pKa = 9.94KK119 pKa = 10.82NLLHH123 pKa = 6.32NVWGTCYY130 pKa = 10.35CFKK133 pKa = 10.85CYY135 pKa = 10.1ILWFGLEE142 pKa = 4.02HH143 pKa = 6.21SWIIFLSWKK152 pKa = 10.33GIIANTPYY160 pKa = 10.65RR161 pKa = 11.84CLDD164 pKa = 3.47LL165 pKa = 6.4
Molecular weight: 19.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1532 |
165 |
674 |
383.0 |
43.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.136 ± 1.7 | 2.285 ± 0.732 |
5.091 ± 0.42 | 7.768 ± 0.727 |
5.222 ± 0.902 | 5.091 ± 0.705 |
2.023 ± 0.432 | 4.308 ± 0.208 |
6.593 ± 1.218 | 10.64 ± 0.77 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.807 ± 0.521 | 5.222 ± 0.291 |
5.679 ± 0.962 | 3.982 ± 0.384 |
4.504 ± 0.209 | 6.854 ± 0.287 |
5.875 ± 0.367 | 5.679 ± 0.967 |
1.371 ± 0.314 | 2.872 ± 0.469 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
