
Sanxia tombus-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.94
Get precalculated fractions of proteins
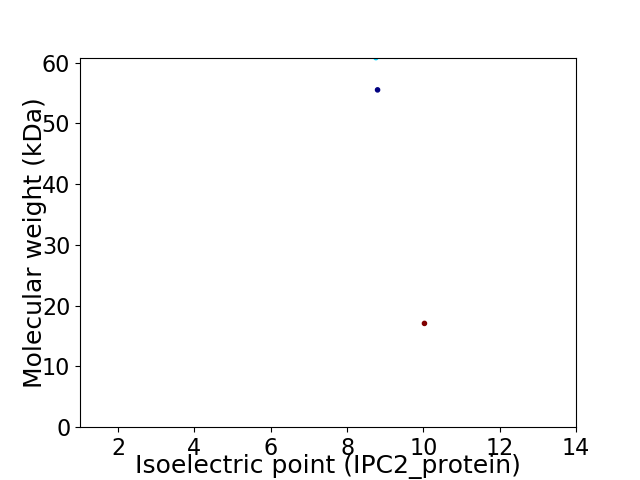
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGD9|A0A1L3KGD9_9VIRU Uncharacterized protein OS=Sanxia tombus-like virus 1 OX=1923385 PE=4 SV=1
MM1 pKa = 7.75DD2 pKa = 3.54MWRR5 pKa = 11.84ILRR8 pKa = 11.84RR9 pKa = 11.84LCFVRR14 pKa = 11.84RR15 pKa = 11.84NRR17 pKa = 11.84CILQMATKK25 pKa = 9.86WLGNLNLLCSRR36 pKa = 11.84ISIVDD41 pKa = 3.91VISLGRR47 pKa = 11.84KK48 pKa = 7.33YY49 pKa = 9.86LTYY52 pKa = 10.81GVLVQGKK59 pKa = 8.89VGQPQLVTFLSTRR72 pKa = 11.84LFIRR76 pKa = 11.84PFQTNIVLHH85 pKa = 5.45TCRR88 pKa = 11.84KK89 pKa = 9.19LLLSSRR95 pKa = 11.84LGLQCYY101 pKa = 9.12PKK103 pKa = 10.68GFNLKK108 pKa = 9.84FVAQNPEE115 pKa = 3.69HH116 pKa = 5.98ATVFGKK122 pKa = 10.66LLEE125 pKa = 4.2QLPSSNWFLNNILARR140 pKa = 11.84FKK142 pKa = 11.1LGINNPVLAICSKK155 pKa = 9.48VSPSIGLSGPEE166 pKa = 3.56KK167 pKa = 10.89VIIKK171 pKa = 8.16MAKK174 pKa = 8.33TKK176 pKa = 10.51NKK178 pKa = 10.32NMTQKK183 pKa = 10.68VGATAMGYY191 pKa = 9.52SNSYY195 pKa = 8.05KK196 pKa = 8.67TPKK199 pKa = 8.38MQTVGSSIIVKK210 pKa = 9.82HH211 pKa = 6.25KK212 pKa = 10.18EE213 pKa = 3.85LIRR216 pKa = 11.84QVASQSAFNVAEE228 pKa = 3.86ARR230 pKa = 11.84INPADD235 pKa = 4.04LEE237 pKa = 4.51SFPWLAGIATRR248 pKa = 11.84YY249 pKa = 7.85EE250 pKa = 4.09KK251 pKa = 11.14YY252 pKa = 10.13RR253 pKa = 11.84FLKK256 pKa = 10.21MKK258 pKa = 8.96FTLVPQVPTTAPGSLGLYY276 pKa = 9.95FDD278 pKa = 5.13YY279 pKa = 11.21DD280 pKa = 3.88ATDD283 pKa = 3.84LPAPNATQFFSNLNAVTTQIWMEE306 pKa = 3.98ASTVVLPQPMEE317 pKa = 4.2LYY319 pKa = 10.71CATKK323 pKa = 10.75YY324 pKa = 10.69DD325 pKa = 3.76GQNPKK330 pKa = 9.32WFDD333 pKa = 3.38YY334 pKa = 11.2GRR336 pKa = 11.84IKK338 pKa = 11.09YY339 pKa = 9.47FMQSSVQCLGYY350 pKa = 10.79LFVEE354 pKa = 4.71YY355 pKa = 9.95EE356 pKa = 4.24VEE358 pKa = 3.89LSKK361 pKa = 10.44PAQTVGQISRR371 pKa = 11.84FSGHH375 pKa = 7.17AYY377 pKa = 8.05WQTLSQSADD386 pKa = 3.48VPVGTPTWIEE396 pKa = 3.79AVTVDD401 pKa = 4.47GAPGQISVPYY411 pKa = 8.74SGWYY415 pKa = 8.85RR416 pKa = 11.84VQAICNSTVNTATLIEE432 pKa = 4.27ICGSAIAALPLGLDD446 pKa = 3.34ANTNPKK452 pKa = 8.99TYY454 pKa = 9.91AYY456 pKa = 8.44TAIVFLTANVPFYY469 pKa = 10.95AFKK472 pKa = 10.94VSAGFVDD479 pKa = 4.37VTADD483 pKa = 3.57PNGADD488 pKa = 3.34SPLGIEE494 pKa = 4.59LEE496 pKa = 4.41LVGPVV501 pKa = 3.02
MM1 pKa = 7.75DD2 pKa = 3.54MWRR5 pKa = 11.84ILRR8 pKa = 11.84RR9 pKa = 11.84LCFVRR14 pKa = 11.84RR15 pKa = 11.84NRR17 pKa = 11.84CILQMATKK25 pKa = 9.86WLGNLNLLCSRR36 pKa = 11.84ISIVDD41 pKa = 3.91VISLGRR47 pKa = 11.84KK48 pKa = 7.33YY49 pKa = 9.86LTYY52 pKa = 10.81GVLVQGKK59 pKa = 8.89VGQPQLVTFLSTRR72 pKa = 11.84LFIRR76 pKa = 11.84PFQTNIVLHH85 pKa = 5.45TCRR88 pKa = 11.84KK89 pKa = 9.19LLLSSRR95 pKa = 11.84LGLQCYY101 pKa = 9.12PKK103 pKa = 10.68GFNLKK108 pKa = 9.84FVAQNPEE115 pKa = 3.69HH116 pKa = 5.98ATVFGKK122 pKa = 10.66LLEE125 pKa = 4.2QLPSSNWFLNNILARR140 pKa = 11.84FKK142 pKa = 11.1LGINNPVLAICSKK155 pKa = 9.48VSPSIGLSGPEE166 pKa = 3.56KK167 pKa = 10.89VIIKK171 pKa = 8.16MAKK174 pKa = 8.33TKK176 pKa = 10.51NKK178 pKa = 10.32NMTQKK183 pKa = 10.68VGATAMGYY191 pKa = 9.52SNSYY195 pKa = 8.05KK196 pKa = 8.67TPKK199 pKa = 8.38MQTVGSSIIVKK210 pKa = 9.82HH211 pKa = 6.25KK212 pKa = 10.18EE213 pKa = 3.85LIRR216 pKa = 11.84QVASQSAFNVAEE228 pKa = 3.86ARR230 pKa = 11.84INPADD235 pKa = 4.04LEE237 pKa = 4.51SFPWLAGIATRR248 pKa = 11.84YY249 pKa = 7.85EE250 pKa = 4.09KK251 pKa = 11.14YY252 pKa = 10.13RR253 pKa = 11.84FLKK256 pKa = 10.21MKK258 pKa = 8.96FTLVPQVPTTAPGSLGLYY276 pKa = 9.95FDD278 pKa = 5.13YY279 pKa = 11.21DD280 pKa = 3.88ATDD283 pKa = 3.84LPAPNATQFFSNLNAVTTQIWMEE306 pKa = 3.98ASTVVLPQPMEE317 pKa = 4.2LYY319 pKa = 10.71CATKK323 pKa = 10.75YY324 pKa = 10.69DD325 pKa = 3.76GQNPKK330 pKa = 9.32WFDD333 pKa = 3.38YY334 pKa = 11.2GRR336 pKa = 11.84IKK338 pKa = 11.09YY339 pKa = 9.47FMQSSVQCLGYY350 pKa = 10.79LFVEE354 pKa = 4.71YY355 pKa = 9.95EE356 pKa = 4.24VEE358 pKa = 3.89LSKK361 pKa = 10.44PAQTVGQISRR371 pKa = 11.84FSGHH375 pKa = 7.17AYY377 pKa = 8.05WQTLSQSADD386 pKa = 3.48VPVGTPTWIEE396 pKa = 3.79AVTVDD401 pKa = 4.47GAPGQISVPYY411 pKa = 8.74SGWYY415 pKa = 8.85RR416 pKa = 11.84VQAICNSTVNTATLIEE432 pKa = 4.27ICGSAIAALPLGLDD446 pKa = 3.34ANTNPKK452 pKa = 8.99TYY454 pKa = 9.91AYY456 pKa = 8.44TAIVFLTANVPFYY469 pKa = 10.95AFKK472 pKa = 10.94VSAGFVDD479 pKa = 4.37VTADD483 pKa = 3.57PNGADD488 pKa = 3.34SPLGIEE494 pKa = 4.59LEE496 pKa = 4.41LVGPVV501 pKa = 3.02
Molecular weight: 55.52 kDa
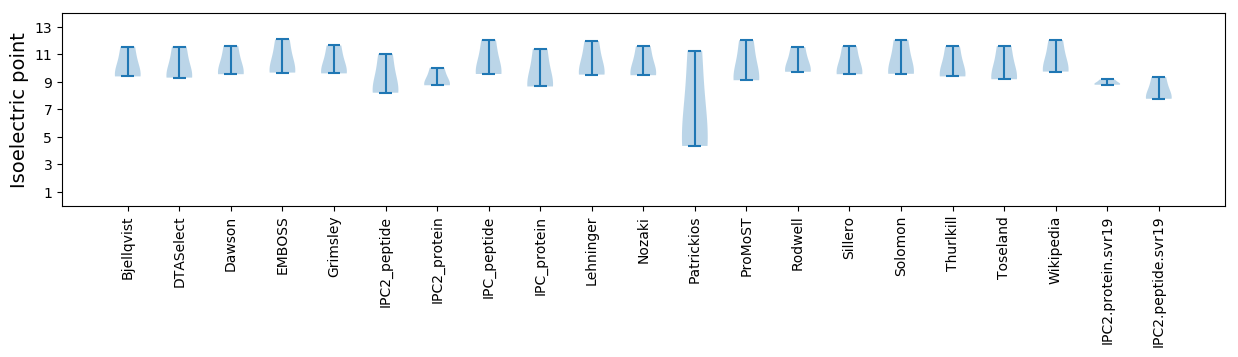
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGZ3|A0A1L3KGZ3_9VIRU RNA-directed RNA polymerase OS=Sanxia tombus-like virus 1 OX=1923385 PE=4 SV=1
MM1 pKa = 7.36CRR3 pKa = 11.84CKK5 pKa = 10.47KK6 pKa = 10.34RR7 pKa = 11.84LLLRR11 pKa = 11.84KK12 pKa = 8.98MNGSSKK18 pKa = 10.57SRR20 pKa = 11.84VVHH23 pKa = 5.35ATLLARR29 pKa = 11.84QLLQKK34 pKa = 10.4QQVTLINITTGRR46 pKa = 11.84VLVILRR52 pKa = 11.84LTGWFLGTLFARR64 pKa = 11.84LSKK67 pKa = 10.47SSKK70 pKa = 9.51FRR72 pKa = 11.84SGALTCVVLWIRR84 pKa = 11.84PSMRR88 pKa = 11.84SCTFHH93 pKa = 8.16GIICIALRR101 pKa = 11.84LGVVVNKK108 pKa = 9.91WKK110 pKa = 10.47DD111 pKa = 3.08SRR113 pKa = 11.84LRR115 pKa = 11.84SRR117 pKa = 11.84LDD119 pKa = 3.13SQIFRR124 pKa = 11.84DD125 pKa = 4.22SHH127 pKa = 6.75SNWPNQLAVSPYY139 pKa = 10.49AVGWDD144 pKa = 3.26SWLAACC150 pKa = 6.05
MM1 pKa = 7.36CRR3 pKa = 11.84CKK5 pKa = 10.47KK6 pKa = 10.34RR7 pKa = 11.84LLLRR11 pKa = 11.84KK12 pKa = 8.98MNGSSKK18 pKa = 10.57SRR20 pKa = 11.84VVHH23 pKa = 5.35ATLLARR29 pKa = 11.84QLLQKK34 pKa = 10.4QQVTLINITTGRR46 pKa = 11.84VLVILRR52 pKa = 11.84LTGWFLGTLFARR64 pKa = 11.84LSKK67 pKa = 10.47SSKK70 pKa = 9.51FRR72 pKa = 11.84SGALTCVVLWIRR84 pKa = 11.84PSMRR88 pKa = 11.84SCTFHH93 pKa = 8.16GIICIALRR101 pKa = 11.84LGVVVNKK108 pKa = 9.91WKK110 pKa = 10.47DD111 pKa = 3.08SRR113 pKa = 11.84LRR115 pKa = 11.84SRR117 pKa = 11.84LDD119 pKa = 3.13SQIFRR124 pKa = 11.84DD125 pKa = 4.22SHH127 pKa = 6.75SNWPNQLAVSPYY139 pKa = 10.49AVGWDD144 pKa = 3.26SWLAACC150 pKa = 6.05
Molecular weight: 17.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1178 |
150 |
527 |
392.7 |
44.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.112 ± 1.009 | 2.377 ± 0.48 |
3.48 ± 0.504 | 3.565 ± 1.162 |
4.669 ± 0.394 | 6.367 ± 0.313 |
1.868 ± 0.571 | 4.924 ± 0.52 |
5.603 ± 0.119 | 11.036 ± 1.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.547 ± 0.287 | 4.244 ± 0.511 |
4.669 ± 0.921 | 6.112 ± 1.006 |
6.452 ± 1.572 | 7.555 ± 0.912 |
5.348 ± 0.89 | 7.64 ± 0.343 |
2.037 ± 0.576 | 3.396 ± 0.823 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
