
Cytophagaceae bacterium BCCC1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; unclassified Cytophagaceae
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
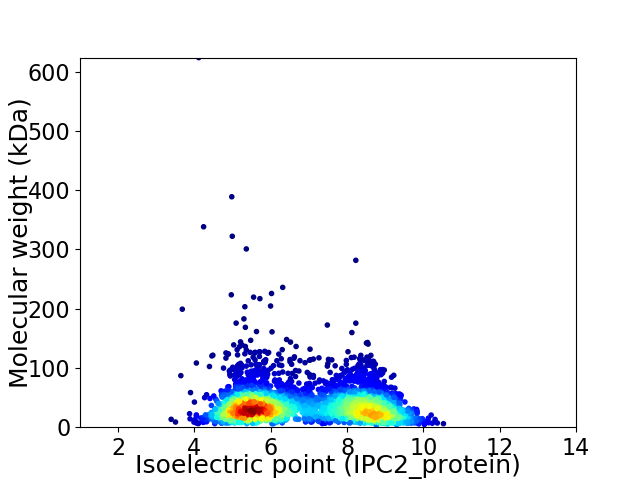
Virtual 2D-PAGE plot for 3514 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A257ISG9|A0A257ISG9_9BACT Uncharacterized protein OS=Cytophagaceae bacterium BCCC1 OX=2015573 GN=CFE22_12280 PE=4 SV=1
MM1 pKa = 7.33TSIAFNGFAEE11 pKa = 4.84GTKK14 pKa = 10.01QVSPTDD20 pKa = 3.36QKK22 pKa = 11.26VASLYY27 pKa = 8.88FAPTNAFGSGFNALQRR43 pKa = 11.84NRR45 pKa = 11.84IYY47 pKa = 10.79FHH49 pKa = 6.84IANNATEE56 pKa = 4.0NLYY59 pKa = 10.88FGANLRR65 pKa = 11.84TGGSTSTLTDD75 pKa = 2.87VYY77 pKa = 11.49YY78 pKa = 10.5RR79 pKa = 11.84IKK81 pKa = 10.68DD82 pKa = 3.26AAGNVVSGPTLFTSNITSHH101 pKa = 6.08AQAVAGPSSYY111 pKa = 11.56SPITFNPTSDD121 pKa = 2.9GDD123 pKa = 3.96FYY125 pKa = 11.33IEE127 pKa = 4.2IYY129 pKa = 10.11RR130 pKa = 11.84GNSGGSAQGSTTAFVAPWFDD150 pKa = 3.73FSVGTSTTAITDD162 pKa = 3.13GRR164 pKa = 11.84VFCEE168 pKa = 3.51NWSFRR173 pKa = 11.84ATKK176 pKa = 10.21PSDD179 pKa = 3.29NFTGTFTDD187 pKa = 4.14PVSPTLYY194 pKa = 10.48SYY196 pKa = 11.4TNDD199 pKa = 3.17EE200 pKa = 4.36TVVKK204 pKa = 11.04VEE206 pKa = 4.62FNDD209 pKa = 4.02FQPLSFIPAFNSYY222 pKa = 10.42GVDD225 pKa = 3.44PLQTDD230 pKa = 3.33WSIGRR235 pKa = 11.84KK236 pKa = 9.28SINSNVSAPVLSGGYY251 pKa = 8.18RR252 pKa = 11.84TFLNEE257 pKa = 3.86PDD259 pKa = 3.35ASVYY263 pKa = 10.47PSASLAAAPTLNGNVTGCAGAYY285 pKa = 7.53FIPYY289 pKa = 7.2FTNVDD294 pKa = 2.84GDD296 pKa = 3.82VRR298 pKa = 11.84FLLDD302 pKa = 3.21INGTAGFQLGTTDD315 pKa = 5.19RR316 pKa = 11.84ILEE319 pKa = 4.26AFGVSAGSHH328 pKa = 5.66LMPWDD333 pKa = 3.93GLDD336 pKa = 3.3GLGNVLSPNTTVTSQISMLRR356 pKa = 11.84GRR358 pKa = 11.84INLPMYY364 pKa = 9.85DD365 pKa = 3.68AEE367 pKa = 4.32MNRR370 pKa = 11.84NGFSLTAVSPVLVSNLRR387 pKa = 11.84MYY389 pKa = 10.41WDD391 pKa = 4.4DD392 pKa = 4.56ASLTNHH398 pKa = 6.72ANNTTGAGVDD408 pKa = 3.72NSALGQVSPGHH419 pKa = 5.98GWNGTYY425 pKa = 10.68GSTLLVPPVLPGGEE439 pKa = 4.44GSDD442 pKa = 3.84TPSDD446 pKa = 3.85ANDD449 pKa = 3.5DD450 pKa = 4.03FGNLRR455 pKa = 11.84TINTWFWGLEE465 pKa = 4.15EE466 pKa = 4.85QSGTLNFRR474 pKa = 11.84LPYY477 pKa = 10.0CISIQGNVYY486 pKa = 10.82SDD488 pKa = 4.14ANGLNNSLINGTGSNFGGLYY508 pKa = 10.2AYY510 pKa = 10.34LVNTLGNVVAIDD522 pKa = 3.88TVNSDD527 pKa = 2.66GTYY530 pKa = 9.98IFNNIDD536 pKa = 3.31SGSFTVRR543 pKa = 11.84ISTNTGVVGNPAPVIAFPTGYY564 pKa = 8.46STIGEE569 pKa = 4.21GTAASGDD576 pKa = 3.83GTPSGDD582 pKa = 3.45TPITVVASNITGVNFGLNQLPNSNTASGTFLNPTGTNTVTVPTLGGSDD630 pKa = 4.32PEE632 pKa = 5.37DD633 pKa = 3.97GSLGVTNSIIIKK645 pKa = 8.95VLPLNGTLYY654 pKa = 10.26YY655 pKa = 10.85NGIAVVDD662 pKa = 4.05EE663 pKa = 4.08QVIASYY669 pKa = 11.34NSALLTLDD677 pKa = 3.54PSFPSVGTVVFDD689 pKa = 3.7YY690 pKa = 11.53AFFDD694 pKa = 3.5NTGLVDD700 pKa = 3.76PTPATVAINFTNNPPVANNDD720 pKa = 3.11TNAPIASTAGQTAINALTATDD741 pKa = 3.71TDD743 pKa = 4.04GTIANYY749 pKa = 10.2RR750 pKa = 11.84IEE752 pKa = 4.19TLPTLGTLYY761 pKa = 10.76VGGVAATVGQVLTPAQVATLTYY783 pKa = 10.41DD784 pKa = 3.39PSGTANGTDD793 pKa = 3.11SFTFTAIDD801 pKa = 3.41NNGAEE806 pKa = 4.73DD807 pKa = 3.71ATDD810 pKa = 3.42ATVSIPVTNNPPVANNDD827 pKa = 3.11TNAPIASTAGQTAINALTATDD848 pKa = 3.71TDD850 pKa = 4.04GTIANYY856 pKa = 10.15RR857 pKa = 11.84IEE859 pKa = 4.25SLPTLGTLYY868 pKa = 10.76VGGVAATVGQVLTPAQVATLTYY890 pKa = 10.41DD891 pKa = 3.39PSGTANGTDD900 pKa = 3.11SFTFTAIDD908 pKa = 3.41NNGAEE913 pKa = 4.73DD914 pKa = 3.73ATDD917 pKa = 3.46ATISIPVTNNPPVANNNTNAPIASTAGQTAINALTATDD955 pKa = 3.71TDD957 pKa = 4.04GTIANYY963 pKa = 10.15RR964 pKa = 11.84IEE966 pKa = 4.46SLPTMGTLYY975 pKa = 10.76VGGVAATVGQVLTPAQVATLTYY997 pKa = 10.41DD998 pKa = 3.39PSGTANGTDD1007 pKa = 3.11SFTFTAIDD1015 pKa = 3.41NNGAEE1020 pKa = 4.73DD1021 pKa = 3.71ATDD1024 pKa = 3.42ATVSIPVNNNPPVANNNTNAPIASTAGQTAINALTATDD1062 pKa = 3.71TDD1064 pKa = 4.04GTIANYY1070 pKa = 10.15RR1071 pKa = 11.84IEE1073 pKa = 4.46SLPTMGTLYY1082 pKa = 10.76VGGVAATVGQVLTPAQVATLTYY1104 pKa = 10.73DD1105 pKa = 3.47PAGTANGTDD1114 pKa = 3.23SFTFTAIDD1122 pKa = 3.41NNGAEE1127 pKa = 4.73DD1128 pKa = 3.73ATDD1131 pKa = 3.46ATISIPVNNIPPIALDD1147 pKa = 5.64DD1148 pKa = 4.12SDD1150 pKa = 5.41LYY1152 pKa = 11.6NAFGTNGIVNILANDD1167 pKa = 3.95TLSTGILATTLNANIDD1183 pKa = 4.25LNPATPALDD1192 pKa = 3.22NTLIVGGEE1200 pKa = 3.88GTYY1203 pKa = 9.92TYY1205 pKa = 10.9NLSNGQVTFVPEE1217 pKa = 3.69IGLTGNPAPINYY1229 pKa = 9.33VLIDD1233 pKa = 3.79NNTGQKK1239 pKa = 8.76DD1240 pKa = 3.39TASIVITYY1248 pKa = 10.33SGFPNAVNDD1257 pKa = 3.7EE1258 pKa = 4.45DD1259 pKa = 5.48LNNLPGIVGVVNALSNDD1276 pKa = 3.76TLSTGAIANTTNASIDD1292 pKa = 4.26LNPLTPTVDD1301 pKa = 3.83HH1302 pKa = 7.09ILTISGQGTYY1312 pKa = 9.93QYY1314 pKa = 11.33NLTTGIVTFIPDD1326 pKa = 3.39NGLIGNPTPINYY1338 pKa = 9.6NLIEE1342 pKa = 4.25LTSGFTDD1349 pKa = 3.24MATISMTYY1357 pKa = 9.82LLPVVAVNDD1366 pKa = 4.3TNTSSPVGVNVTFDD1380 pKa = 3.14IVSNDD1385 pKa = 3.33SLSNGAIATPAEE1397 pKa = 4.12VLVDD1401 pKa = 3.84INLTTPGYY1409 pKa = 9.47QDD1411 pKa = 3.37SLIVSGEE1418 pKa = 4.34GIWSYY1423 pKa = 11.84NSTTGEE1429 pKa = 3.78ITFDD1433 pKa = 3.7PEE1435 pKa = 3.96TGFVNDD1441 pKa = 4.65PTPISYY1447 pKa = 10.24IITEE1451 pKa = 4.28AMTGLLDD1458 pKa = 3.58TATISIMYY1466 pKa = 7.78GVPPIANSDD1475 pKa = 3.37ADD1477 pKa = 5.7LINQPQTSTVINLLANDD1494 pKa = 4.63LLSNLSTATFSMVDD1508 pKa = 3.18VDD1510 pKa = 5.92LEE1512 pKa = 4.2PTALGIQTSVLVPGEE1527 pKa = 3.88GNYY1530 pKa = 10.1VYY1532 pKa = 10.93NSSNGNLTFTPSIGNYY1548 pKa = 10.09SNPTPLIYY1556 pKa = 10.66SLIEE1560 pKa = 4.03KK1561 pKa = 9.09ATGLSDD1567 pKa = 3.43TARR1570 pKa = 11.84VTIQYY1575 pKa = 9.37QFSPSLEE1582 pKa = 4.29LIKK1585 pKa = 10.29TNTLMGSGLVGDD1597 pKa = 4.41SVQYY1601 pKa = 11.14DD1602 pKa = 3.18FTVINTGNVPVTGITITDD1620 pKa = 3.69TKK1622 pKa = 10.51ISSTPIPVTPSALAPGGTAVASRR1645 pKa = 11.84MYY1647 pKa = 10.56YY1648 pKa = 8.99LTAGDD1653 pKa = 3.65ILAGEE1658 pKa = 4.61VLNSALVNGTSSNAIAVKK1676 pKa = 9.36DD1677 pKa = 3.69TSDD1680 pKa = 3.42NGDD1683 pKa = 3.29PGMPGRR1689 pKa = 11.84SNPTRR1694 pKa = 11.84LLLPLSSDD1702 pKa = 3.07VDD1704 pKa = 3.8VKK1706 pKa = 10.86LTGTIIGNCEE1716 pKa = 3.86RR1717 pKa = 11.84VVGDD1721 pKa = 3.37IVTYY1725 pKa = 6.92QVKK1728 pKa = 9.27VFRR1731 pKa = 11.84EE1732 pKa = 3.95DD1733 pKa = 3.33SNAGLVDD1740 pKa = 3.7VVVKK1744 pKa = 10.87DD1745 pKa = 3.54SLGVNFEE1752 pKa = 4.54LVSQIATEE1760 pKa = 4.38GSFNIGTSRR1769 pKa = 11.84WSGISLASGDD1779 pKa = 3.87TATLTFQLRR1788 pKa = 11.84ILTNIGGLTCVEE1800 pKa = 4.28SWVDD1804 pKa = 3.36SLSRR1808 pKa = 11.84ADD1810 pKa = 4.57LDD1812 pKa = 4.28SNPGNRR1818 pKa = 11.84ITTEE1822 pKa = 3.68DD1823 pKa = 4.48DD1824 pKa = 2.69IARR1827 pKa = 11.84ACVSVPISICTTKK1840 pKa = 11.07AEE1842 pKa = 4.17TAEE1845 pKa = 4.38LSTSLGYY1852 pKa = 10.28SSYY1855 pKa = 9.28QWYY1858 pKa = 10.48RR1859 pKa = 11.84NGTLISGAISPTFTATQGGSYY1880 pKa = 8.86TVIVNGSLCPNGSCCPIVVQEE1901 pKa = 4.18NCPCPVEE1908 pKa = 3.96ICLPIAIRR1916 pKa = 11.84KK1917 pKa = 6.22TRR1919 pKa = 3.2
MM1 pKa = 7.33TSIAFNGFAEE11 pKa = 4.84GTKK14 pKa = 10.01QVSPTDD20 pKa = 3.36QKK22 pKa = 11.26VASLYY27 pKa = 8.88FAPTNAFGSGFNALQRR43 pKa = 11.84NRR45 pKa = 11.84IYY47 pKa = 10.79FHH49 pKa = 6.84IANNATEE56 pKa = 4.0NLYY59 pKa = 10.88FGANLRR65 pKa = 11.84TGGSTSTLTDD75 pKa = 2.87VYY77 pKa = 11.49YY78 pKa = 10.5RR79 pKa = 11.84IKK81 pKa = 10.68DD82 pKa = 3.26AAGNVVSGPTLFTSNITSHH101 pKa = 6.08AQAVAGPSSYY111 pKa = 11.56SPITFNPTSDD121 pKa = 2.9GDD123 pKa = 3.96FYY125 pKa = 11.33IEE127 pKa = 4.2IYY129 pKa = 10.11RR130 pKa = 11.84GNSGGSAQGSTTAFVAPWFDD150 pKa = 3.73FSVGTSTTAITDD162 pKa = 3.13GRR164 pKa = 11.84VFCEE168 pKa = 3.51NWSFRR173 pKa = 11.84ATKK176 pKa = 10.21PSDD179 pKa = 3.29NFTGTFTDD187 pKa = 4.14PVSPTLYY194 pKa = 10.48SYY196 pKa = 11.4TNDD199 pKa = 3.17EE200 pKa = 4.36TVVKK204 pKa = 11.04VEE206 pKa = 4.62FNDD209 pKa = 4.02FQPLSFIPAFNSYY222 pKa = 10.42GVDD225 pKa = 3.44PLQTDD230 pKa = 3.33WSIGRR235 pKa = 11.84KK236 pKa = 9.28SINSNVSAPVLSGGYY251 pKa = 8.18RR252 pKa = 11.84TFLNEE257 pKa = 3.86PDD259 pKa = 3.35ASVYY263 pKa = 10.47PSASLAAAPTLNGNVTGCAGAYY285 pKa = 7.53FIPYY289 pKa = 7.2FTNVDD294 pKa = 2.84GDD296 pKa = 3.82VRR298 pKa = 11.84FLLDD302 pKa = 3.21INGTAGFQLGTTDD315 pKa = 5.19RR316 pKa = 11.84ILEE319 pKa = 4.26AFGVSAGSHH328 pKa = 5.66LMPWDD333 pKa = 3.93GLDD336 pKa = 3.3GLGNVLSPNTTVTSQISMLRR356 pKa = 11.84GRR358 pKa = 11.84INLPMYY364 pKa = 9.85DD365 pKa = 3.68AEE367 pKa = 4.32MNRR370 pKa = 11.84NGFSLTAVSPVLVSNLRR387 pKa = 11.84MYY389 pKa = 10.41WDD391 pKa = 4.4DD392 pKa = 4.56ASLTNHH398 pKa = 6.72ANNTTGAGVDD408 pKa = 3.72NSALGQVSPGHH419 pKa = 5.98GWNGTYY425 pKa = 10.68GSTLLVPPVLPGGEE439 pKa = 4.44GSDD442 pKa = 3.84TPSDD446 pKa = 3.85ANDD449 pKa = 3.5DD450 pKa = 4.03FGNLRR455 pKa = 11.84TINTWFWGLEE465 pKa = 4.15EE466 pKa = 4.85QSGTLNFRR474 pKa = 11.84LPYY477 pKa = 10.0CISIQGNVYY486 pKa = 10.82SDD488 pKa = 4.14ANGLNNSLINGTGSNFGGLYY508 pKa = 10.2AYY510 pKa = 10.34LVNTLGNVVAIDD522 pKa = 3.88TVNSDD527 pKa = 2.66GTYY530 pKa = 9.98IFNNIDD536 pKa = 3.31SGSFTVRR543 pKa = 11.84ISTNTGVVGNPAPVIAFPTGYY564 pKa = 8.46STIGEE569 pKa = 4.21GTAASGDD576 pKa = 3.83GTPSGDD582 pKa = 3.45TPITVVASNITGVNFGLNQLPNSNTASGTFLNPTGTNTVTVPTLGGSDD630 pKa = 4.32PEE632 pKa = 5.37DD633 pKa = 3.97GSLGVTNSIIIKK645 pKa = 8.95VLPLNGTLYY654 pKa = 10.26YY655 pKa = 10.85NGIAVVDD662 pKa = 4.05EE663 pKa = 4.08QVIASYY669 pKa = 11.34NSALLTLDD677 pKa = 3.54PSFPSVGTVVFDD689 pKa = 3.7YY690 pKa = 11.53AFFDD694 pKa = 3.5NTGLVDD700 pKa = 3.76PTPATVAINFTNNPPVANNDD720 pKa = 3.11TNAPIASTAGQTAINALTATDD741 pKa = 3.71TDD743 pKa = 4.04GTIANYY749 pKa = 10.2RR750 pKa = 11.84IEE752 pKa = 4.19TLPTLGTLYY761 pKa = 10.76VGGVAATVGQVLTPAQVATLTYY783 pKa = 10.41DD784 pKa = 3.39PSGTANGTDD793 pKa = 3.11SFTFTAIDD801 pKa = 3.41NNGAEE806 pKa = 4.73DD807 pKa = 3.71ATDD810 pKa = 3.42ATVSIPVTNNPPVANNDD827 pKa = 3.11TNAPIASTAGQTAINALTATDD848 pKa = 3.71TDD850 pKa = 4.04GTIANYY856 pKa = 10.15RR857 pKa = 11.84IEE859 pKa = 4.25SLPTLGTLYY868 pKa = 10.76VGGVAATVGQVLTPAQVATLTYY890 pKa = 10.41DD891 pKa = 3.39PSGTANGTDD900 pKa = 3.11SFTFTAIDD908 pKa = 3.41NNGAEE913 pKa = 4.73DD914 pKa = 3.73ATDD917 pKa = 3.46ATISIPVTNNPPVANNNTNAPIASTAGQTAINALTATDD955 pKa = 3.71TDD957 pKa = 4.04GTIANYY963 pKa = 10.15RR964 pKa = 11.84IEE966 pKa = 4.46SLPTMGTLYY975 pKa = 10.76VGGVAATVGQVLTPAQVATLTYY997 pKa = 10.41DD998 pKa = 3.39PSGTANGTDD1007 pKa = 3.11SFTFTAIDD1015 pKa = 3.41NNGAEE1020 pKa = 4.73DD1021 pKa = 3.71ATDD1024 pKa = 3.42ATVSIPVNNNPPVANNNTNAPIASTAGQTAINALTATDD1062 pKa = 3.71TDD1064 pKa = 4.04GTIANYY1070 pKa = 10.15RR1071 pKa = 11.84IEE1073 pKa = 4.46SLPTMGTLYY1082 pKa = 10.76VGGVAATVGQVLTPAQVATLTYY1104 pKa = 10.73DD1105 pKa = 3.47PAGTANGTDD1114 pKa = 3.23SFTFTAIDD1122 pKa = 3.41NNGAEE1127 pKa = 4.73DD1128 pKa = 3.73ATDD1131 pKa = 3.46ATISIPVNNIPPIALDD1147 pKa = 5.64DD1148 pKa = 4.12SDD1150 pKa = 5.41LYY1152 pKa = 11.6NAFGTNGIVNILANDD1167 pKa = 3.95TLSTGILATTLNANIDD1183 pKa = 4.25LNPATPALDD1192 pKa = 3.22NTLIVGGEE1200 pKa = 3.88GTYY1203 pKa = 9.92TYY1205 pKa = 10.9NLSNGQVTFVPEE1217 pKa = 3.69IGLTGNPAPINYY1229 pKa = 9.33VLIDD1233 pKa = 3.79NNTGQKK1239 pKa = 8.76DD1240 pKa = 3.39TASIVITYY1248 pKa = 10.33SGFPNAVNDD1257 pKa = 3.7EE1258 pKa = 4.45DD1259 pKa = 5.48LNNLPGIVGVVNALSNDD1276 pKa = 3.76TLSTGAIANTTNASIDD1292 pKa = 4.26LNPLTPTVDD1301 pKa = 3.83HH1302 pKa = 7.09ILTISGQGTYY1312 pKa = 9.93QYY1314 pKa = 11.33NLTTGIVTFIPDD1326 pKa = 3.39NGLIGNPTPINYY1338 pKa = 9.6NLIEE1342 pKa = 4.25LTSGFTDD1349 pKa = 3.24MATISMTYY1357 pKa = 9.82LLPVVAVNDD1366 pKa = 4.3TNTSSPVGVNVTFDD1380 pKa = 3.14IVSNDD1385 pKa = 3.33SLSNGAIATPAEE1397 pKa = 4.12VLVDD1401 pKa = 3.84INLTTPGYY1409 pKa = 9.47QDD1411 pKa = 3.37SLIVSGEE1418 pKa = 4.34GIWSYY1423 pKa = 11.84NSTTGEE1429 pKa = 3.78ITFDD1433 pKa = 3.7PEE1435 pKa = 3.96TGFVNDD1441 pKa = 4.65PTPISYY1447 pKa = 10.24IITEE1451 pKa = 4.28AMTGLLDD1458 pKa = 3.58TATISIMYY1466 pKa = 7.78GVPPIANSDD1475 pKa = 3.37ADD1477 pKa = 5.7LINQPQTSTVINLLANDD1494 pKa = 4.63LLSNLSTATFSMVDD1508 pKa = 3.18VDD1510 pKa = 5.92LEE1512 pKa = 4.2PTALGIQTSVLVPGEE1527 pKa = 3.88GNYY1530 pKa = 10.1VYY1532 pKa = 10.93NSSNGNLTFTPSIGNYY1548 pKa = 10.09SNPTPLIYY1556 pKa = 10.66SLIEE1560 pKa = 4.03KK1561 pKa = 9.09ATGLSDD1567 pKa = 3.43TARR1570 pKa = 11.84VTIQYY1575 pKa = 9.37QFSPSLEE1582 pKa = 4.29LIKK1585 pKa = 10.29TNTLMGSGLVGDD1597 pKa = 4.41SVQYY1601 pKa = 11.14DD1602 pKa = 3.18FTVINTGNVPVTGITITDD1620 pKa = 3.69TKK1622 pKa = 10.51ISSTPIPVTPSALAPGGTAVASRR1645 pKa = 11.84MYY1647 pKa = 10.56YY1648 pKa = 8.99LTAGDD1653 pKa = 3.65ILAGEE1658 pKa = 4.61VLNSALVNGTSSNAIAVKK1676 pKa = 9.36DD1677 pKa = 3.69TSDD1680 pKa = 3.42NGDD1683 pKa = 3.29PGMPGRR1689 pKa = 11.84SNPTRR1694 pKa = 11.84LLLPLSSDD1702 pKa = 3.07VDD1704 pKa = 3.8VKK1706 pKa = 10.86LTGTIIGNCEE1716 pKa = 3.86RR1717 pKa = 11.84VVGDD1721 pKa = 3.37IVTYY1725 pKa = 6.92QVKK1728 pKa = 9.27VFRR1731 pKa = 11.84EE1732 pKa = 3.95DD1733 pKa = 3.33SNAGLVDD1740 pKa = 3.7VVVKK1744 pKa = 10.87DD1745 pKa = 3.54SLGVNFEE1752 pKa = 4.54LVSQIATEE1760 pKa = 4.38GSFNIGTSRR1769 pKa = 11.84WSGISLASGDD1779 pKa = 3.87TATLTFQLRR1788 pKa = 11.84ILTNIGGLTCVEE1800 pKa = 4.28SWVDD1804 pKa = 3.36SLSRR1808 pKa = 11.84ADD1810 pKa = 4.57LDD1812 pKa = 4.28SNPGNRR1818 pKa = 11.84ITTEE1822 pKa = 3.68DD1823 pKa = 4.48DD1824 pKa = 2.69IARR1827 pKa = 11.84ACVSVPISICTTKK1840 pKa = 11.07AEE1842 pKa = 4.17TAEE1845 pKa = 4.38LSTSLGYY1852 pKa = 10.28SSYY1855 pKa = 9.28QWYY1858 pKa = 10.48RR1859 pKa = 11.84NGTLISGAISPTFTATQGGSYY1880 pKa = 8.86TVIVNGSLCPNGSCCPIVVQEE1901 pKa = 4.18NCPCPVEE1908 pKa = 3.96ICLPIAIRR1916 pKa = 11.84KK1917 pKa = 6.22TRR1919 pKa = 3.2
Molecular weight: 199.1 kDa
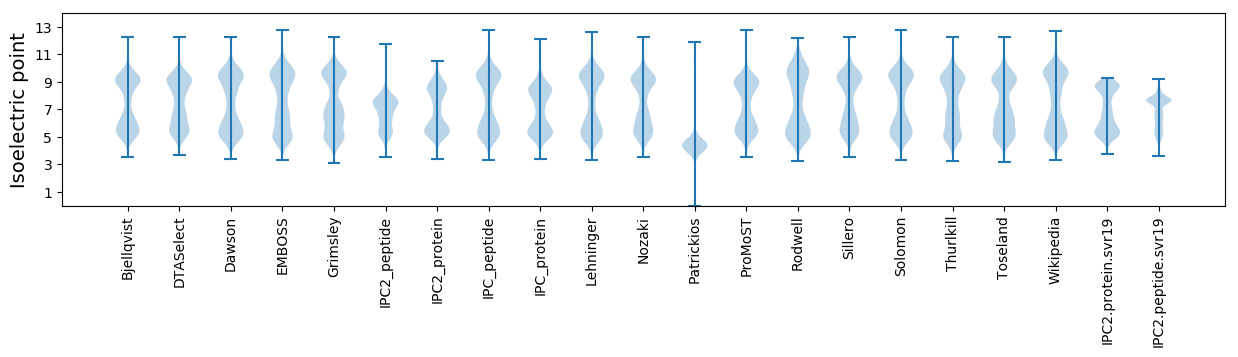
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A257J1M5|A0A257J1M5_9BACT PnuC protein OS=Cytophagaceae bacterium BCCC1 OX=2015573 GN=CFE22_00745 PE=4 SV=1
MM1 pKa = 7.41AQLCSRR7 pKa = 11.84GRR9 pKa = 11.84YY10 pKa = 8.42FDD12 pKa = 3.61KK13 pKa = 11.02KK14 pKa = 10.74SFFLGFVRR22 pKa = 11.84HH23 pKa = 5.87PRR25 pKa = 11.84FNGGVYY31 pKa = 9.53TYY33 pKa = 10.77TSFFEE38 pKa = 4.25KK39 pKa = 10.25TSVNTLRR46 pKa = 11.84RR47 pKa = 11.84HH48 pKa = 4.76WNGVNTGVNRR58 pKa = 11.84GVNILYY64 pKa = 9.43TGAA67 pKa = 3.96
MM1 pKa = 7.41AQLCSRR7 pKa = 11.84GRR9 pKa = 11.84YY10 pKa = 8.42FDD12 pKa = 3.61KK13 pKa = 11.02KK14 pKa = 10.74SFFLGFVRR22 pKa = 11.84HH23 pKa = 5.87PRR25 pKa = 11.84FNGGVYY31 pKa = 9.53TYY33 pKa = 10.77TSFFEE38 pKa = 4.25KK39 pKa = 10.25TSVNTLRR46 pKa = 11.84RR47 pKa = 11.84HH48 pKa = 4.76WNGVNTGVNRR58 pKa = 11.84GVNILYY64 pKa = 9.43TGAA67 pKa = 3.96
Molecular weight: 7.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1214332 |
30 |
6045 |
345.6 |
38.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.447 ± 0.044 | 0.839 ± 0.019 |
5.116 ± 0.028 | 6.28 ± 0.057 |
5.578 ± 0.044 | 6.808 ± 0.045 |
1.633 ± 0.019 | 7.848 ± 0.044 |
8.012 ± 0.065 | 9.311 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.297 ± 0.024 | 6.151 ± 0.043 |
3.622 ± 0.026 | 3.282 ± 0.024 |
3.29 ± 0.026 | 6.811 ± 0.048 |
5.576 ± 0.084 | 6.106 ± 0.037 |
1.162 ± 0.015 | 3.832 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
