
Reinekea forsetii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Saccharospirillaceae; Reinekea
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
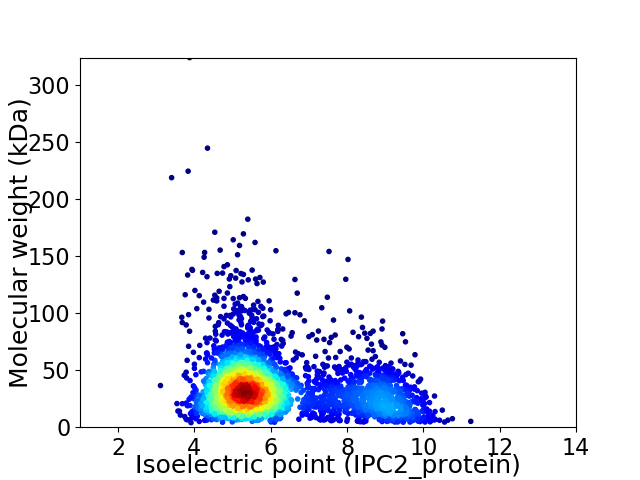
Virtual 2D-PAGE plot for 3259 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K8KN44|A0A2K8KN44_9GAMM Proteorhodopsin OS=Reinekea forsetii OX=1336806 GN=REIFOR_00373 PE=3 SV=1
MM1 pKa = 7.54ASIGVLLGCAQPAGSDD17 pKa = 3.37TSGTTDD23 pKa = 2.95GTADD27 pKa = 3.19ATIYY31 pKa = 10.3RR32 pKa = 11.84QIAGTSALGRR42 pKa = 11.84AGSWSSNCLGSDD54 pKa = 4.02LASEE58 pKa = 4.75CARR61 pKa = 11.84ADD63 pKa = 3.55SEE65 pKa = 4.78GNYY68 pKa = 10.43LLSTPAEE75 pKa = 4.19QSQLMWSDD83 pKa = 3.12IPEE86 pKa = 4.31SDD88 pKa = 3.13GTTVRR93 pKa = 11.84LYY95 pKa = 10.94SRR97 pKa = 11.84YY98 pKa = 9.02RR99 pKa = 11.84HH100 pKa = 5.62QDD102 pKa = 2.78GLTSALININPSTHH116 pKa = 6.26AVLDD120 pKa = 3.33IWSNSQQGQSLEE132 pKa = 4.2LCAEE136 pKa = 4.03EE137 pKa = 4.44ATCIDD142 pKa = 3.67ALLAGFSAAVEE153 pKa = 4.11QTIVAQLDD161 pKa = 3.85EE162 pKa = 4.9LLGEE166 pKa = 4.66AWPDD170 pKa = 3.25GRR172 pKa = 11.84DD173 pKa = 3.65PFDD176 pKa = 4.43DD177 pKa = 4.75VYY179 pKa = 11.5LADD182 pKa = 5.39PDD184 pKa = 5.08LDD186 pKa = 4.23EE187 pKa = 6.45LDD189 pKa = 4.96LMHH192 pKa = 7.23DD193 pKa = 3.44TLGFVVSSSDD203 pKa = 4.33LSVIDD208 pKa = 3.61NAGEE212 pKa = 3.96ILTQVSLGRR221 pKa = 11.84LLQAVEE227 pKa = 4.3LADD230 pKa = 3.81LTLTSAQISAAQALPIPDD248 pKa = 4.21PEE250 pKa = 5.95IIVISLRR257 pKa = 11.84MSVSPTLATEE267 pKa = 4.15APVDD271 pKa = 3.65VLVSDD276 pKa = 4.74RR277 pKa = 11.84GSSSVAGGEE286 pKa = 4.19LTFSHH291 pKa = 7.7DD292 pKa = 3.23LTLANGASLSFSGADD307 pKa = 3.08VVTTITAGGDD317 pKa = 3.68HH318 pKa = 6.48NWVVTATDD326 pKa = 3.6INDD329 pKa = 3.46FSITQGYY336 pKa = 8.99VISVLNGSDD345 pKa = 3.44GVATFGGAGSCLTPLTGLNANALNQCHH372 pKa = 5.54EE373 pKa = 4.49VQNGTALGSCDD384 pKa = 3.61QLDD387 pKa = 3.88SGSVSLSDD395 pKa = 4.54SPAPCARR402 pKa = 11.84EE403 pKa = 3.98VQNEE407 pKa = 3.94GALFGVCTWVDD418 pKa = 3.06SEE420 pKa = 4.73LRR422 pKa = 11.84VFHH425 pKa = 6.18YY426 pKa = 8.87EE427 pKa = 3.63NPLRR431 pKa = 11.84LNLVEE436 pKa = 5.23LFSEE440 pKa = 4.33RR441 pKa = 11.84QSRR444 pKa = 11.84IATEE448 pKa = 4.61CISDD452 pKa = 3.88VGNLWSISPP461 pKa = 3.95
MM1 pKa = 7.54ASIGVLLGCAQPAGSDD17 pKa = 3.37TSGTTDD23 pKa = 2.95GTADD27 pKa = 3.19ATIYY31 pKa = 10.3RR32 pKa = 11.84QIAGTSALGRR42 pKa = 11.84AGSWSSNCLGSDD54 pKa = 4.02LASEE58 pKa = 4.75CARR61 pKa = 11.84ADD63 pKa = 3.55SEE65 pKa = 4.78GNYY68 pKa = 10.43LLSTPAEE75 pKa = 4.19QSQLMWSDD83 pKa = 3.12IPEE86 pKa = 4.31SDD88 pKa = 3.13GTTVRR93 pKa = 11.84LYY95 pKa = 10.94SRR97 pKa = 11.84YY98 pKa = 9.02RR99 pKa = 11.84HH100 pKa = 5.62QDD102 pKa = 2.78GLTSALININPSTHH116 pKa = 6.26AVLDD120 pKa = 3.33IWSNSQQGQSLEE132 pKa = 4.2LCAEE136 pKa = 4.03EE137 pKa = 4.44ATCIDD142 pKa = 3.67ALLAGFSAAVEE153 pKa = 4.11QTIVAQLDD161 pKa = 3.85EE162 pKa = 4.9LLGEE166 pKa = 4.66AWPDD170 pKa = 3.25GRR172 pKa = 11.84DD173 pKa = 3.65PFDD176 pKa = 4.43DD177 pKa = 4.75VYY179 pKa = 11.5LADD182 pKa = 5.39PDD184 pKa = 5.08LDD186 pKa = 4.23EE187 pKa = 6.45LDD189 pKa = 4.96LMHH192 pKa = 7.23DD193 pKa = 3.44TLGFVVSSSDD203 pKa = 4.33LSVIDD208 pKa = 3.61NAGEE212 pKa = 3.96ILTQVSLGRR221 pKa = 11.84LLQAVEE227 pKa = 4.3LADD230 pKa = 3.81LTLTSAQISAAQALPIPDD248 pKa = 4.21PEE250 pKa = 5.95IIVISLRR257 pKa = 11.84MSVSPTLATEE267 pKa = 4.15APVDD271 pKa = 3.65VLVSDD276 pKa = 4.74RR277 pKa = 11.84GSSSVAGGEE286 pKa = 4.19LTFSHH291 pKa = 7.7DD292 pKa = 3.23LTLANGASLSFSGADD307 pKa = 3.08VVTTITAGGDD317 pKa = 3.68HH318 pKa = 6.48NWVVTATDD326 pKa = 3.6INDD329 pKa = 3.46FSITQGYY336 pKa = 8.99VISVLNGSDD345 pKa = 3.44GVATFGGAGSCLTPLTGLNANALNQCHH372 pKa = 5.54EE373 pKa = 4.49VQNGTALGSCDD384 pKa = 3.61QLDD387 pKa = 3.88SGSVSLSDD395 pKa = 4.54SPAPCARR402 pKa = 11.84EE403 pKa = 3.98VQNEE407 pKa = 3.94GALFGVCTWVDD418 pKa = 3.06SEE420 pKa = 4.73LRR422 pKa = 11.84VFHH425 pKa = 6.18YY426 pKa = 8.87EE427 pKa = 3.63NPLRR431 pKa = 11.84LNLVEE436 pKa = 5.23LFSEE440 pKa = 4.33RR441 pKa = 11.84QSRR444 pKa = 11.84IATEE448 pKa = 4.61CISDD452 pKa = 3.88VGNLWSISPP461 pKa = 3.95
Molecular weight: 48.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K8KVS9|A0A2K8KVS9_9GAMM Kinase OS=Reinekea forsetii OX=1336806 GN=REIFOR_02728 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.01RR12 pKa = 11.84ARR14 pKa = 11.84SHH16 pKa = 6.1GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.43NGRR28 pKa = 11.84KK29 pKa = 9.65LIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84ALLSAA44 pKa = 4.86
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.01RR12 pKa = 11.84ARR14 pKa = 11.84SHH16 pKa = 6.1GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.43NGRR28 pKa = 11.84KK29 pKa = 9.65LIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84ALLSAA44 pKa = 4.86
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1072189 |
37 |
3058 |
329.0 |
36.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.1 ± 0.064 | 0.944 ± 0.014 |
5.734 ± 0.041 | 5.395 ± 0.042 |
3.974 ± 0.032 | 7.211 ± 0.049 |
2.219 ± 0.021 | 5.929 ± 0.034 |
3.825 ± 0.034 | 11.23 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.464 ± 0.02 | 3.658 ± 0.026 |
4.262 ± 0.028 | 4.799 ± 0.036 |
5.414 ± 0.037 | 6.354 ± 0.035 |
5.342 ± 0.037 | 6.99 ± 0.032 |
1.371 ± 0.019 | 2.784 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
