
Pseudozyma antarctica (Yeast) (Candida antarctica)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Ustilaginomycotina; Ustilaginomycetes; Ustilaginales; Ustilaginaceae; Moesziomyces
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4672 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A081CKH7|A0A081CKH7_PSEA2 Probable Shwachman-Bodian-Diamond syndrome protein OS=Pseudozyma antarctica OX=84753 GN=PAN0_017c5399 PE=3 SV=1
MM1 pKa = 7.37PVSSASNLVPGASVSLLGFASLAFVVHH28 pKa = 6.47CALFEE33 pKa = 4.4GEE35 pKa = 4.51FLRR38 pKa = 11.84LIAVSAMWSVLVATPLCICAAAAQCIDD65 pKa = 3.69CARR68 pKa = 11.84AAGCTDD74 pKa = 3.45CHH76 pKa = 6.25QAFDD80 pKa = 4.09ARR82 pKa = 11.84ASNSSADD89 pKa = 3.36FDD91 pKa = 4.18RR92 pKa = 11.84PEE94 pKa = 3.68RR95 pKa = 11.84TYY97 pKa = 10.83PVAVKK102 pKa = 9.02TLNLAGFFDD111 pKa = 5.14CLDD114 pKa = 3.91APDD117 pKa = 4.52YY118 pKa = 11.09PSDD121 pKa = 3.7QIIYY125 pKa = 10.09AVDD128 pKa = 3.41RR129 pKa = 11.84QGVFIEE135 pKa = 4.12VDD137 pKa = 2.98MGDD140 pKa = 3.93LLEE143 pKa = 4.35HH144 pKa = 6.79ARR146 pKa = 11.84RR147 pKa = 11.84PTQDD151 pKa = 3.37CNHH154 pKa = 5.53VVTDD158 pKa = 3.57EE159 pKa = 4.41TTVLDD164 pKa = 4.29YY165 pKa = 10.39LTLFHH170 pKa = 6.87GPSLGGRR177 pKa = 11.84SVALAAEE184 pKa = 4.76DD185 pKa = 3.67EE186 pKa = 4.87TIVGSNIDD194 pKa = 4.46PDD196 pKa = 3.91QSDD199 pKa = 4.31PDD201 pKa = 4.53PDD203 pKa = 4.0TVLLGQPIGPEE214 pKa = 4.23FWAARR219 pKa = 11.84HH220 pKa = 5.66GADD223 pKa = 4.04FCAEE227 pKa = 3.86
MM1 pKa = 7.37PVSSASNLVPGASVSLLGFASLAFVVHH28 pKa = 6.47CALFEE33 pKa = 4.4GEE35 pKa = 4.51FLRR38 pKa = 11.84LIAVSAMWSVLVATPLCICAAAAQCIDD65 pKa = 3.69CARR68 pKa = 11.84AAGCTDD74 pKa = 3.45CHH76 pKa = 6.25QAFDD80 pKa = 4.09ARR82 pKa = 11.84ASNSSADD89 pKa = 3.36FDD91 pKa = 4.18RR92 pKa = 11.84PEE94 pKa = 3.68RR95 pKa = 11.84TYY97 pKa = 10.83PVAVKK102 pKa = 9.02TLNLAGFFDD111 pKa = 5.14CLDD114 pKa = 3.91APDD117 pKa = 4.52YY118 pKa = 11.09PSDD121 pKa = 3.7QIIYY125 pKa = 10.09AVDD128 pKa = 3.41RR129 pKa = 11.84QGVFIEE135 pKa = 4.12VDD137 pKa = 2.98MGDD140 pKa = 3.93LLEE143 pKa = 4.35HH144 pKa = 6.79ARR146 pKa = 11.84RR147 pKa = 11.84PTQDD151 pKa = 3.37CNHH154 pKa = 5.53VVTDD158 pKa = 3.57EE159 pKa = 4.41TTVLDD164 pKa = 4.29YY165 pKa = 10.39LTLFHH170 pKa = 6.87GPSLGGRR177 pKa = 11.84SVALAAEE184 pKa = 4.76DD185 pKa = 3.67EE186 pKa = 4.87TIVGSNIDD194 pKa = 4.46PDD196 pKa = 3.91QSDD199 pKa = 4.31PDD201 pKa = 4.53PDD203 pKa = 4.0TVLLGQPIGPEE214 pKa = 4.23FWAARR219 pKa = 11.84HH220 pKa = 5.66GADD223 pKa = 4.04FCAEE227 pKa = 3.86
Molecular weight: 24.18 kDa
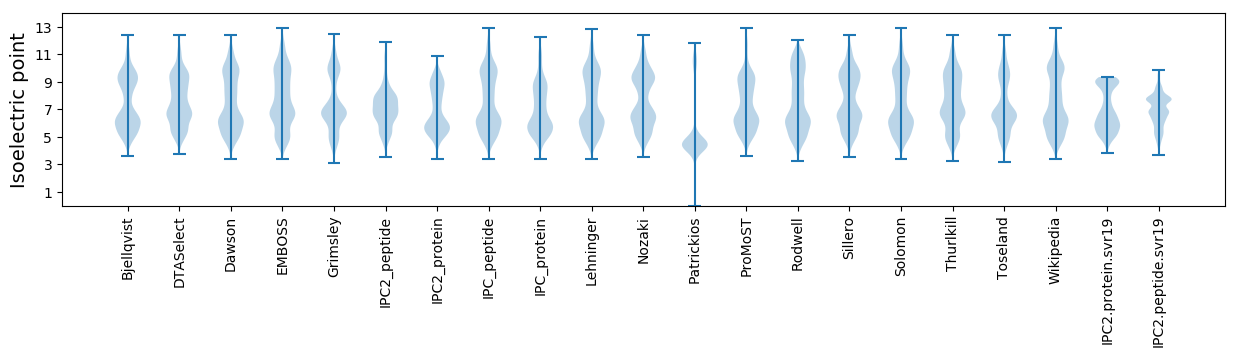
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A081CMQ8|A0A081CMQ8_PSEA2 D-ribose-5-phosphate ketol-isomerase OS=Pseudozyma antarctica OX=84753 GN=PAN0_028c6184 PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 10.05RR3 pKa = 11.84KK4 pKa = 9.37KK5 pKa = 9.97MKK7 pKa = 8.83MKK9 pKa = 10.77LMILMMVTLLLLLQCNDD26 pKa = 3.33QPEE29 pKa = 4.26LADD32 pKa = 3.67RR33 pKa = 11.84VSVVPVLSICDD44 pKa = 3.98ALEE47 pKa = 4.1HH48 pKa = 6.62AATSLPHH55 pKa = 6.01KK56 pKa = 9.58HH57 pKa = 6.1AKK59 pKa = 7.69KK60 pKa = 8.9TLRR63 pKa = 11.84LDD65 pKa = 3.4RR66 pKa = 11.84KK67 pKa = 8.49TPLPCCAAALLPPALLRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84SASLILARR94 pKa = 11.84SGSSLLGVDD103 pKa = 3.49HH104 pKa = 7.21RR105 pKa = 11.84LVISPSSTAAQAACAPARR123 pKa = 11.84AALHH127 pKa = 5.85LAHH130 pKa = 7.18RR131 pKa = 11.84PLLASFPAPVVRR143 pKa = 11.84ISARR147 pKa = 11.84DD148 pKa = 3.45GLDD151 pKa = 3.29PLTASVSLEE160 pKa = 3.69SRR162 pKa = 11.84RR163 pKa = 11.84FRR165 pKa = 11.84PYY167 pKa = 9.67MRR169 pKa = 11.84EE170 pKa = 3.29SCYY173 pKa = 10.46RR174 pKa = 11.84IARR177 pKa = 11.84RR178 pKa = 11.84SSGPYY183 pKa = 9.44QISEE187 pKa = 4.16VSLAWSFASNDD198 pKa = 3.59TSLSTPACLLRR209 pKa = 11.84AFPYY213 pKa = 9.08STSSLLVISSVTKK226 pKa = 10.61RR227 pKa = 11.84SS228 pKa = 3.2
MM1 pKa = 7.28KK2 pKa = 10.05RR3 pKa = 11.84KK4 pKa = 9.37KK5 pKa = 9.97MKK7 pKa = 8.83MKK9 pKa = 10.77LMILMMVTLLLLLQCNDD26 pKa = 3.33QPEE29 pKa = 4.26LADD32 pKa = 3.67RR33 pKa = 11.84VSVVPVLSICDD44 pKa = 3.98ALEE47 pKa = 4.1HH48 pKa = 6.62AATSLPHH55 pKa = 6.01KK56 pKa = 9.58HH57 pKa = 6.1AKK59 pKa = 7.69KK60 pKa = 8.9TLRR63 pKa = 11.84LDD65 pKa = 3.4RR66 pKa = 11.84KK67 pKa = 8.49TPLPCCAAALLPPALLRR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84SASLILARR94 pKa = 11.84SGSSLLGVDD103 pKa = 3.49HH104 pKa = 7.21RR105 pKa = 11.84LVISPSSTAAQAACAPARR123 pKa = 11.84AALHH127 pKa = 5.85LAHH130 pKa = 7.18RR131 pKa = 11.84PLLASFPAPVVRR143 pKa = 11.84ISARR147 pKa = 11.84DD148 pKa = 3.45GLDD151 pKa = 3.29PLTASVSLEE160 pKa = 3.69SRR162 pKa = 11.84RR163 pKa = 11.84FRR165 pKa = 11.84PYY167 pKa = 9.67MRR169 pKa = 11.84EE170 pKa = 3.29SCYY173 pKa = 10.46RR174 pKa = 11.84IARR177 pKa = 11.84RR178 pKa = 11.84SSGPYY183 pKa = 9.44QISEE187 pKa = 4.16VSLAWSFASNDD198 pKa = 3.59TSLSTPACLLRR209 pKa = 11.84AFPYY213 pKa = 9.08STSSLLVISSVTKK226 pKa = 10.61RR227 pKa = 11.84SS228 pKa = 3.2
Molecular weight: 24.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2859900 |
33 |
5427 |
612.1 |
66.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.786 ± 0.042 | 0.989 ± 0.01 |
5.804 ± 0.028 | 5.339 ± 0.027 |
3.152 ± 0.017 | 6.777 ± 0.03 |
2.643 ± 0.016 | 3.781 ± 0.023 |
4.104 ± 0.028 | 8.834 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.931 ± 0.01 | 2.992 ± 0.017 |
6.468 ± 0.038 | 4.198 ± 0.03 |
6.708 ± 0.027 | 9.532 ± 0.053 |
5.737 ± 0.022 | 5.877 ± 0.027 |
1.192 ± 0.012 | 2.153 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
